
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,556,046 – 7,556,159 |
| Length | 113 |
| Max. P | 0.718444 |

| Location | 7,556,046 – 7,556,159 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.40 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -12.18 |
| Energy contribution | -13.90 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
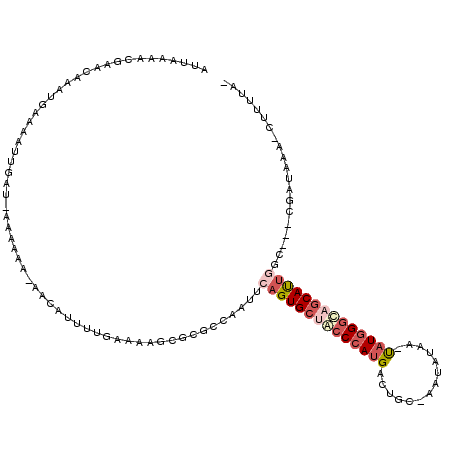
>X_DroMel_CAF1 7556046 113 + 22224390 AUUAAAAGGAACAAAUGAAAAUUGAU--AAAGA-AAUAUUUUGAACGGCGCGCCAAUUAAGUGCUACCCAUGCGGGC-AAUUUAA-CAUGGCUAGCAUUGACCGUCGAUAAA-UUUUUA- ...............(((((((((((--(....-..)))).....(((((.(.((.....((((((.(((((.....-.......-))))).)))))))).))))))...))-))))))- ( -19.90) >DroSec_CAF1 48548 117 + 1 AUUAAAAGGAACAAAUGAAAAUUGAUUAAAAAA-AAUAUUUUGAACGGGGCGCCAAUUCUGUGCUACCCAUGAAGAC-AAUAUAAGUAUGGGUAGCAUUGGCCGUCGAUAAUCUUUUUA- ..((((((((...............((((((..-....)))))).(((.(.((((.....((((((((((((.....-........))))))))))))))))).)))....))))))))- ( -31.92) >DroEre_CAF1 59924 117 + 1 AUUAAAACAAACAAAUGAAAUUUGAUAAAAAAA-AACAUUUUGAAAAUCGCGCCAAUUAAGUGCGACCCAUGCCUCCGAAUGUACGUAUGGGCAGCAUUGGCUAACGAUUAA-CUUUUA- ...........(((((...))))).........-...........(((((.((((.....((((..(((((((............)))))))..))))))))...)))))..-......- ( -23.60) >DroYak_CAF1 50034 100 + 1 AUUAAAACCAACAAAUGCAAAUUGAUAAAAAUG-GACAUUUUGAAAUGCGCGCCAAUUCAGUGCGACCCAUGACUGCUAAUAUACAUAUGGGCAGCAUUGG------------------- ................((..(((....(((((.-...)))))..)))..)).......((((((..((((((..((........))))))))..)))))).------------------- ( -19.00) >DroAna_CAF1 51463 93 + 1 UUUAA--------AUUCAAAUUUA---AACGCGGAAGAACCGGAAGAUGGCGCCCAAACAGUGCUGCCCAUGUCUG----------UCU-GGCAACACUGCC----AGUUGG-CCGUUAC (((((--------((....)))))---)).((((..((..((((....(((((.(.....).)).)))....))))----------.((-((((....))))----))))..-))))... ( -23.90) >consensus AUUAAAACGAACAAAUGAAAAUUGAU_AAAAAA_AACAUUUUGAAAAGCGCGCCAAUUCAGUGCUACCCAUGACUGC_AAUAUAA_UAUGGGCAGCAUUGGC___CGAUAAA_CUUUUA_ ..........................................................((((((((((((((..............)))))))))))))).................... (-12.18 = -13.90 + 1.72)

| Location | 7,556,046 – 7,556,159 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.40 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -12.67 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.623987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7556046 113 - 22224390 -UAAAAA-UUUAUCGACGGUCAAUGCUAGCCAUG-UUAAAUU-GCCCGCAUGGGUAGCACUUAAUUGGCGCGCCGUUCAAAAUAUU-UCUUU--AUCAAUUUUCAUUUGUUCCUUUUAAU -......-.....((.((((((((((((.(((((-(......-....)))))).))))).....))))).)).))...........-.....--.......................... ( -21.20) >DroSec_CAF1 48548 117 - 1 -UAAAAAGAUUAUCGACGGCCAAUGCUACCCAUACUUAUAUU-GUCUUCAUGGGUAGCACAGAAUUGGCGCCCCGUUCAAAAUAUU-UUUUUUAAUCAAUUUUCAUUUGUUCCUUUUAAU -((((((((.(((.(((((((((((((((((((((.......-))....)))))))))).....)))))....))))....))).)-))))))).......................... ( -25.20) >DroEre_CAF1 59924 117 - 1 -UAAAAG-UUAAUCGUUAGCCAAUGCUGCCCAUACGUACAUUCGGAGGCAUGGGUCGCACUUAAUUGGCGCGAUUUUCAAAAUGUU-UUUUUUUAUCAAAUUUCAUUUGUUUGUUUUAAU -((((((-..((((((..((((((((.((((((.(.(........).).)))))).))).....)))))))))))..)........-.........(((((.......)))))))))).. ( -25.00) >DroYak_CAF1 50034 100 - 1 -------------------CCAAUGCUGCCCAUAUGUAUAUUAGCAGUCAUGGGUCGCACUGAAUUGGCGCGCAUUUCAAAAUGUC-CAUUUUUAUCAAUUUGCAUUUGUUGGUUUUAAU -------------------(((((((.((((((.(((......)))...)))))).))..((((.((.....)).))))((((((.-.(((......)))..)))))))))))....... ( -21.40) >DroAna_CAF1 51463 93 - 1 GUAACGG-CCAACU----GGCAGUGUUGCC-AGA----------CAGACAUGGGCAGCACUGUUUGGGCGCCAUCUUCCGGUUCUUCCGCGUU---UAAAUUUGAAU--------UUAAA ..(((((-((....----((((((((((((-...----------........))))))))))))..))).........(((.....)))))))---...........--------..... ( -29.20) >consensus _UAAAAA_UUAAUCG___GCCAAUGCUGCCCAUA_GUAUAUU_GCAGGCAUGGGUAGCACUGAAUUGGCGCGCCGUUCAAAAUAUU_UUUUUU_AUCAAUUUUCAUUUGUUCCUUUUAAU ..................(((((((((((((((................)))))))))).....)))))................................................... (-12.67 = -13.43 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:31 2006