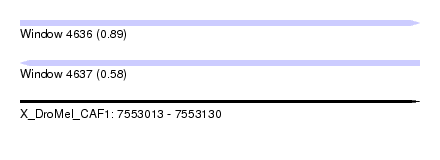
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,553,013 – 7,553,130 |
| Length | 117 |
| Max. P | 0.890698 |

| Location | 7,553,013 – 7,553,130 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.36 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7553013 117 + 22224390 GCGCAUGUUGUUGCUGUGCAGCGCAAACGACGACGACAGCGACAAAUGUGUAGAUCAACAGAUCAGUCACUCGAAAAGCGAAACUAAAAAGUAACCAAAUAAAAAAAAUGAAACA--AA ((((((.(((((((((((..(((....)).)..).)))))))))))))))).((((....))))..(((.(((.....))).(((....)))................)))....--.. ( -25.80) >DroPse_CAF1 107523 101 + 1 ---------GUUGCCGUGCAGAGUAGUCGACUACGACAGGGACAAAUGUGGGGAUCAACAGAUCAUUCACUCAAA-------ACGAAAACGUAACGAAA--AUAAGAAUGAAAGGAAAA ---------(((((.((.(...((((....)))).....).))....(((((((((....)))).))))).....-------........)))))....--.................. ( -18.40) >DroSec_CAF1 45595 116 + 1 GCGCAUGUUGUUGCUGUGCAGCGCAAACGACGACGACAGCGACAAAUGUGUAGAUCAACAGAUCAGUCACUCAAAAAGCGAAACUAAAAAGUAACCAAAUAAA-AAAAUGAAACA--AA ((((((.(((((((((((..(((....)).)..).)))))))))))))))).((((....)))).......................................-...........--.. ( -24.50) >DroSim_CAF1 44899 116 + 1 GCGCAUGUUGUUGCUGUGCAGCGCAAACGACGACGACAGCGACAAAUGUGUAGAUCAACAGAUCAGUCACUCAAAAAGCGAAACUAAAAAGUAACCAAAUAAA-AAAAUGAAACA--AA ((((((.(((((((((((..(((....)).)..).)))))))))))))))).((((....)))).......................................-...........--.. ( -24.50) >DroPer_CAF1 113417 101 + 1 ---------GUUGCCGUACAGAGUGGUCGACUACGACAGGGACAAAUGUGGAGAUCAACAGAUCAUUCACUCAAA-------ACGAAAACGUAACGAAA--AUAAGAAUGAAAGGAAAA ---------((((((((...(((((((((....))))...............((((....))))...)))))...-------))).....)))))....--.................. ( -19.70) >consensus GCGCAUGUUGUUGCUGUGCAGCGCAAACGACGACGACAGCGACAAAUGUGUAGAUCAACAGAUCAGUCACUCAAAAAGCGAAACUAAAAAGUAACCAAAUAAAAAAAAUGAAACA__AA .(((.....((((((((.(..((.......))..)))))))))....(((..((((....))))...))).......)))..(((....)))........................... (-14.22 = -14.36 + 0.14)

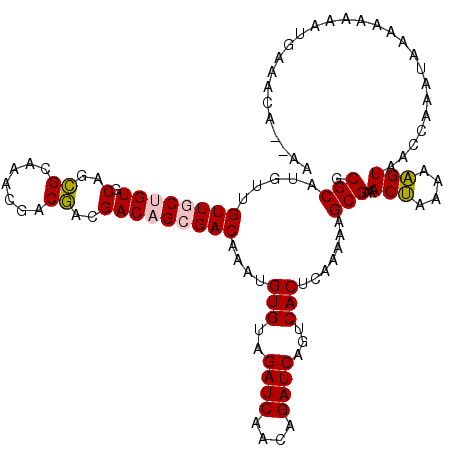
| Location | 7,553,013 – 7,553,130 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -16.18 |
| Energy contribution | -14.90 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7553013 117 - 22224390 UU--UGUUUCAUUUUUUUUAUUUGGUUACUUUUUAGUUUCGCUUUUCGAGUGACUGAUCUGUUGAUCUACACAUUUGUCGCUGUCGUCGUCGUUUGCGCUGCACAGCAACAACAUGCGC ..--...................((((((((...((.....))....))))))))((((....))))..(.(((((((.(((((.((.(.((....))).))))))).)))).))).). ( -25.20) >DroPse_CAF1 107523 101 - 1 UUUUCCUUUCAUUCUUAU--UUUCGUUACGUUUUCGU-------UUUGAGUGAAUGAUCUGUUGAUCCCCACAUUUGUCCCUGUCGUAGUCGACUACUCUGCACGGCAAC--------- ..................--..(((..(((....)))-------..)))(((...((((....))))..)))..(((.((.(((.((((....))))...))).))))).--------- ( -18.30) >DroSec_CAF1 45595 116 - 1 UU--UGUUUCAUUUU-UUUAUUUGGUUACUUUUUAGUUUCGCUUUUUGAGUGACUGAUCUGUUGAUCUACACAUUUGUCGCUGUCGUCGUCGUUUGCGCUGCACAGCAACAACAUGCGC ..--...........-.......((((((((...((.....))....))))))))((((....))))..(.(((((((.(((((.((.(.((....))).))))))).)))).))).). ( -25.90) >DroSim_CAF1 44899 116 - 1 UU--UGUUUCAUUUU-UUUAUUUGGUUACUUUUUAGUUUCGCUUUUUGAGUGACUGAUCUGUUGAUCUACACAUUUGUCGCUGUCGUCGUCGUUUGCGCUGCACAGCAACAACAUGCGC ..--...........-.......((((((((...((.....))....))))))))((((....))))..(.(((((((.(((((.((.(.((....))).))))))).)))).))).). ( -25.90) >DroPer_CAF1 113417 101 - 1 UUUUCCUUUCAUUCUUAU--UUUCGUUACGUUUUCGU-------UUUGAGUGAAUGAUCUGUUGAUCUCCACAUUUGUCCCUGUCGUAGUCGACCACUCUGUACGGCAAC--------- ..................--.........(((.((((-------...(((((...((((....))))...............((((....)))))))))...)))).)))--------- ( -19.30) >consensus UU__UGUUUCAUUUUUUUUAUUUGGUUACUUUUUAGUUUCGCUUUUUGAGUGACUGAUCUGUUGAUCUACACAUUUGUCGCUGUCGUCGUCGUUUGCGCUGCACAGCAACAACAUGCGC ........((((((.............(((....)))..........))))))..((((....)))).........((.(((((((.(((.....))).)).))))).))......... (-16.18 = -14.90 + -1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:27 2006