
| Sequence ID | X_DroMel_CAF1 |
|---|---|
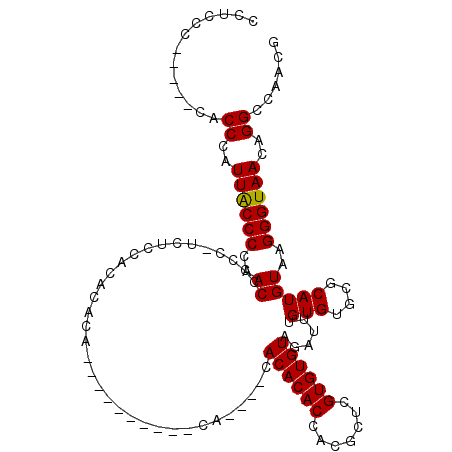
| Location | 7,548,808 – 7,548,910 |
| Length | 102 |
| Max. P | 0.993500 |

| Location | 7,548,808 – 7,548,910 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7548808 102 + 22224390 CCUCCC-----CACCCAUUACCCCAACCCCC-UUUCAACACACACUC----ACACA----CACACACCGCGCUCGUGUGUAGAUUUGUGUGCGCAUGUAAGGGUAACAGGCCAACG ......-----..((..((((((..((....-......((((((.((----(((((----(.............)))))).))..)))))).....))..))))))..))...... ( -21.58) >DroSec_CAF1 41485 110 + 1 AGCGCG-----CACCCAUUACCCCAACUCCC-UUUCCAAACACACUCCUCUGCACAGCGGCACACACCGCGCACGUGUGUAGAUUUGUGUGCGCAUGUAAGGGUAACAGGCCAACG ......-----..........((.....(((-((......(((((...((((((((((((......)))).....))))))))...))))).......))))).....))...... ( -29.82) >DroEre_CAF1 52625 95 + 1 CCUCCC-----CGCCCAUUGCCCCAACCCCCGUCUCCACAAA------------CA----CACACACCACGCUCGUGUGUAGAUUUGUGUGUGCAUGUAAGGGUAACAGGCCAACG ......-----.(((..((((((..((....))....(((..------------((----(((((((.((((....)))).)...))))))))..)))..))))))..)))..... ( -26.50) >DroYak_CAF1 42441 101 + 1 CCUCCCCUACCCACCCAUUACCCCAACCCCC-UCUCCACACACA----------CA----CACACACCACGCUCGUGUGUAGAUUUGUGUGUGCAUGUAAGGGUAACAGGCCAACG .............((..((((((..((....-....((((((((----------..----(((((((.......)))))).)...))))))))...))..))))))..))...... ( -24.80) >consensus CCUCCC_____CACCCAUUACCCCAACCCCC_UCUCCACACACA__________CA____CACACACCACGCUCGUGUGUAGAUUUGUGUGCGCAUGUAAGGGUAACAGGCCAACG .............((..((((((..((..................................((((((.......))))))......(((....)))))..))))))..))...... (-17.01 = -16.82 + -0.19)

| Location | 7,548,808 – 7,548,910 |
|---|---|
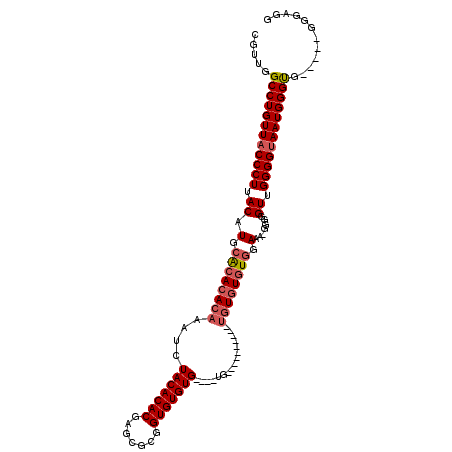
| Length | 102 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -29.66 |
| Energy contribution | -30.73 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7548808 102 - 22224390 CGUUGGCCUGUUACCCUUACAUGCGCACACAAAUCUACACACGAGCGCGGUGUGUG----UGUGU----GAGUGUGUGUUGAAA-GGGGGUUGGGGUAAUGGGUG-----GGGAGG ..((.((((((((((((.((..((((((((..((.((((((((.......))))))----)).))----..)))))))).....-....)).)))))))))))).-----)).... ( -40.80) >DroSec_CAF1 41485 110 - 1 CGUUGGCCUGUUACCCUUACAUGCGCACACAAAUCUACACACGUGCGCGGUGUGUGCCGCUGUGCAGAGGAGUGUGUUUGGAAA-GGGAGUUGGGGUAAUGGGUG-----CGCGCU (((..((((((((((((.((....((((((...(((.((((((.((((.....)))))).)))).)))...))))))((....)-)...)).)))))))))))).-----.))).. ( -43.20) >DroEre_CAF1 52625 95 - 1 CGUUGGCCUGUUACCCUUACAUGCACACACAAAUCUACACACGAGCGUGGUGUGUG----UG------------UUUGUGGAGACGGGGGUUGGGGCAAUGGGCG-----GGGAGG (((((.(((...(((((((((.(((((((((...((((.(....).))))))))))----))------------).)))(....))))))).))).)))))....-----...... ( -38.40) >DroYak_CAF1 42441 101 - 1 CGUUGGCCUGUUACCCUUACAUGCACACACAAAUCUACACACGAGCGUGGUGUGUG----UG----------UGUGUGUGGAGA-GGGGGUUGGGGUAAUGGGUGGGUAGGGGAGG ..((.((((((((((((((((((((((((((...((((.(....).))))..))))----))----------))))))))....-.......)))))))))))).))......... ( -41.60) >consensus CGUUGGCCUGUUACCCUUACAUGCACACACAAAUCUACACACGAGCGCGGUGUGUG____UG__________UGUGUGUGGAAA_GGGGGUUGGGGUAAUGGGUG_____GGGAGG .....((((((((((((.((.(.((((((((....(((((((.......)))))))................)))))))).).......)).))))))))))))............ (-29.66 = -30.73 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:23 2006