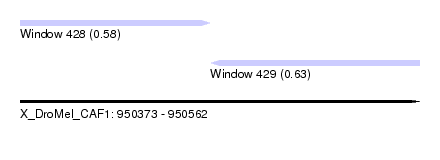
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 950,373 – 950,562 |
| Length | 189 |
| Max. P | 0.630240 |

| Location | 950,373 – 950,463 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 950373 90 + 22224390 AUUUCAGAUUUCU-------GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCAUUUUUACAUUGAUGAAACUUUGCUUCGGCCCCGAAGCCAU----------------------- .....((.((((.-------..((((...(((((((.................)))))))...))))...))))))..((((((....))))))...----------------------- ( -20.03) >DroSec_CAF1 33318 90 + 1 AUUUCUGAUUUCU-------GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCAUUUUUUCAUUGAUGAAACUUUGCUUCGGCCCCGAAGCCAU----------------------- ........((((.-------..((((((.(((((((.................))))))).))))))...))))....((((((....))))))...----------------------- ( -22.13) >DroSim_CAF1 30838 89 + 1 AUUUCAGAUUUCU-------GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCAUUU-UUCAUUGAUGAAACUUUGCUUCGGCCCCGAAGCCAU----------------------- .....((.((((.-------..((((((.(((((((.................)))))))-))))))...))))))..((((((....))))))...----------------------- ( -20.83) >DroEre_CAF1 41631 90 + 1 AUUUCAGAUUUCU-------GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCACUUUUAUAUUGAUGAAACUCUGCUUUGGCCCCGUAGCCAU----------------------- ....((((((((.-------..((((...(((((((.................)))))))...))))...)))).))))...((((......)))).----------------------- ( -19.93) >DroYak_CAF1 35748 113 + 1 AUUUGAGACUUCU-------GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCACUUUUACAUUGAUGAAACUUUGCUUUGGCCCCGAAGCCAUAUUGCCAUAUUGCCAUAUUGCCA .........(((.-------..((((...(((((((.................)))))))...))))...))).....((((((....)))))).((((.((......)).))))..... ( -20.43) >DroAna_CAF1 18854 89 + 1 GGAUCAGAGUUGGGUUGGGGAUGAGGGAUAAGUGCAUUUCAUGAGCCAUAAAUUGCAAUUU-CCAUUCGAGUA-------CUUGGCCCAGGGGCUAC----------------------- .....((..((((((..((..(((((((....((((.((.(((...))).)).))))...)-)).))))....-------))..))))))...))..----------------------- ( -24.10) >consensus AUUUCAGAUUUCU_______GUGAUGGAUAAGUGCAUUUCACAAGCCAUAAAUUGCAUUUUUACAUUGAUGAAACUUUGCUUCGGCCCCGAAGCCAU_______________________ ......................((((...(((((((.................)))))))...))))...........((((((....)))))).......................... (-13.52 = -13.58 + 0.06)


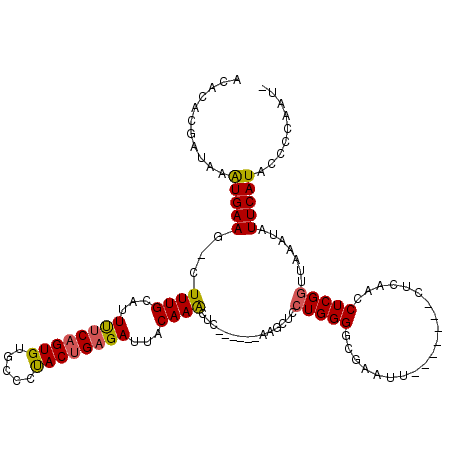
| Location | 950,463 – 950,562 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.01 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 950463 99 - 22224390 ACACACGAUAAAUGAAG-CUUUGCAUUUUCAGUGUGCCCUACUGAGAUUACAAACU------------CCUGGGGCGAAUU-------CUCAACCUCGGUUAAAUAUUCAUACCCCAAA- ..((((...(((((...-.....)))))...))))((((((..(((........))------------).))))))((((.-------...(((....)))....))))..........- ( -22.40) >DroSec_CAF1 33408 111 - 1 ACACACGAUAAAUGAAG-CUUUGCAUUUUCAGUGUGCCCUACUGAGAUUACAAACUCCAGCUCCAGCUCCUGGGGCGAAUU-------CUCAACCUCGGUUAAAUAUUCAUACCCCAAU- .(((((...(((((...-.....)))))...)))))....((((((.............(((((((...)))))))((...-------.))...))))))...................- ( -26.10) >DroSim_CAF1 30927 111 - 1 ACACACGAUAAAUGAAG-CUUUGCAUUUUCAGUGUGCCCUACUGAGAUUACAAACUCCAGCUCCAGCUCCUGGGGCGAAUU-------CUCAACCUCGGUUAAAUAUUCAUACCCCAAU- .(((((...(((((...-.....)))))...)))))....((((((.............(((((((...)))))))((...-------.))...))))))...................- ( -26.10) >DroEre_CAF1 41721 106 - 1 ACACACGAUAAAUGAAAUAUUUGAUUUCUCAGUGUGCUUUACUGAGAUUACAAACUC------AAGCUUCUGGGGCAAAAU-------CUGAAACUCGCUUAAAUAUUCAUACCCCAAU- ...........(((((.(((((((..((((((((.....)))))))).......(((------(......)))).......-------...........))))))))))))........- ( -19.10) >DroYak_CAF1 35861 113 - 1 ACACACGAUAAGUGAAC-CUUUGGGUUUGCAGUGUGCUUCACCGAGAUUACAAGCUC------AAGCUUCUGGGGCAAAUUCUCAAUUCUCAAACUCGGUUAAAUAUUCAUACCCCAAUA .(((((.....(..(((-(....))))..).)))))....((((((.....((((..------..)))).(((((.....))))).........)))))).................... ( -27.00) >consensus ACACACGAUAAAUGAAG_CUUUGCAUUUUCAGUGUGCCCUACUGAGAUUACAAACUC______AAGCUCCUGGGGCGAAUU_______CUCAACCUCGGUUAAAUAUUCAUACCCCAAU_ ...........(((((...((((...((((((((.....))))))))...))))...............(((((....................))))).......)))))......... (-14.05 = -14.01 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:45 2006