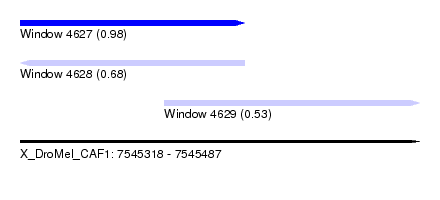
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,545,318 – 7,545,487 |
| Length | 169 |
| Max. P | 0.977074 |

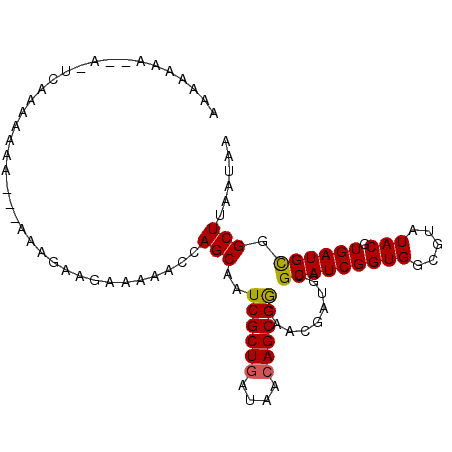
| Location | 7,545,318 – 7,545,413 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -17.81 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7545318 95 + 22224390 AAAAAA---AAUCAAAAAAA---GAAGAAGAAAAACCAGCAAUCGCUGAUAACAGCGGGAACGAUGUGCAUCGGUGGCGUAUACGUGAUGCGGCUUAAUAA ......---...........---..............(((..((((((((.(((.((....)).)))..))))))))(((((.....))))))))...... ( -20.50) >DroSec_CAF1 38402 89 + 1 AAAAGAAA---------AAA---CAAGAAGAAAAACCAGCAAUCGCUGAUAAGAGCGGGAACGAUGUGCAUCGGUGGCGUAUACGUGAUGCGGCUUUAUAA ........---------...---.............((((....))))...((((((....)....((((((((((.....))).)))))))))))).... ( -19.30) >DroEre_CAF1 49320 99 + 1 AAAAACAUAAGUCAAAAAAAUGUAGAGAAGAAAAACCAGCAAUCGCUGAUAACAGCGAGAACGA--UGCAUCGGUGGCGUAUACGUGAUGUGGCUAAAUAA .........(((((.....(((((.....(....(((.(((.((((((....))))))......--)))...)))..)...)))))....)))))...... ( -20.80) >consensus AAAAAAA__A_UCAAAAAAA___AAAGAAGAAAAACCAGCAAUCGCUGAUAACAGCGGGAACGAUGUGCAUCGGUGGCGUAUACGUGAUGCGGCUUAAUAA .....................................(((..((((((....)))))).........(((((((((.....))).)))))).)))...... (-17.81 = -17.70 + -0.11)


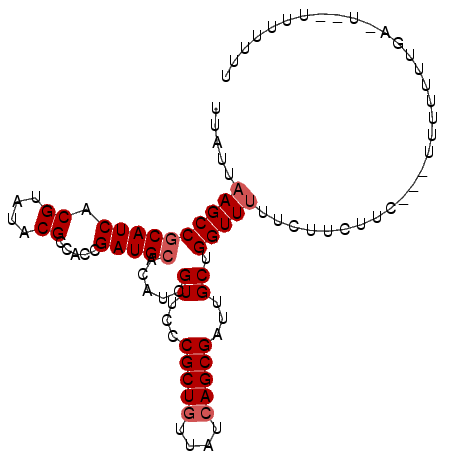
| Location | 7,545,318 – 7,545,413 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -13.27 |
| Energy contribution | -14.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7545318 95 - 22224390 UUAUUAAGCCGCAUCACGUAUACGCCACCGAUGCACAUCGUUCCCGCUGUUAUCAGCGAUUGCUGGUUUUUCUUCUUC---UUUUUUUGAUU---UUUUUU .....((((((((((.((....)).....))))).....((...(((((....)))))...)).))))).........---...........---...... ( -17.80) >DroSec_CAF1 38402 89 - 1 UUAUAAAGCCGCAUCACGUAUACGCCACCGAUGCACAUCGUUCCCGCUCUUAUCAGCGAUUGCUGGUUUUUCUUCUUG---UUU---------UUUCUUUU ....(((((((((((.((....)).....))))).....((...((((......))))...)).))))))........---...---------........ ( -15.40) >DroEre_CAF1 49320 99 - 1 UUAUUUAGCCACAUCACGUAUACGCCACCGAUGCA--UCGUUCUCGCUGUUAUCAGCGAUUGCUGGUUUUUCUUCUCUACAUUUUUUUGACUUAUGUUUUU ......(((((......((((.((....)))))).--..((..((((((....))))))..)))))))................................. ( -17.20) >consensus UUAUUAAGCCGCAUCACGUAUACGCCACCGAUGCACAUCGUUCCCGCUGUUAUCAGCGAUUGCUGGUUUUUCUUCUUC___UUUUUUUGA_U__UUUUUUU .....((((((((((.((....)).....))))).....((...(((((....)))))...)).)))))................................ (-13.27 = -14.27 + 1.00)


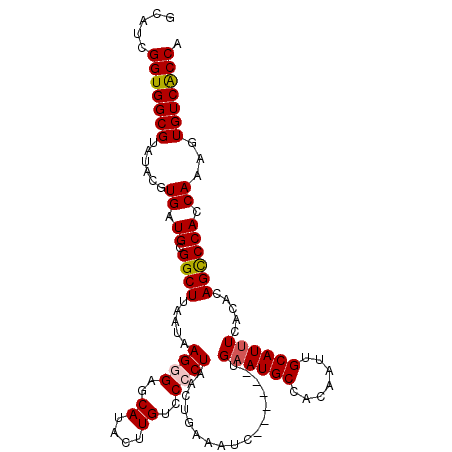
| Location | 7,545,379 – 7,545,487 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 90.94 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -25.88 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7545379 108 + 22224390 GCAUCGGUGGCGUAUACGUGAUGCGGCUUAAUAAGGGAGCAUACUUGUACCCUAACUGAAAUC------UGAAUGCCACAAUUGCAUCUCACACAGCCCACCAAAGUGUCACCA .....(((((((......((.((.((((.....((((.(((....))).))))..........------(((((((.......)))).)))...)))))).))...))))))). ( -28.90) >DroSec_CAF1 38457 108 + 1 GCAUCGGUGGCGUAUACGUGAUGCGGCUUUAUAAGGGAGCAUACUUGUCCCCUAAAUGAAAUC------UGAAUGCCACAAUUGCAUUUCACACAGCCCACCAAAGUGUCACCA .....(((((((......((.((.((((.....((((..((....))..))))..........------.((((((.......)))))).....)))))).))...))))))). ( -29.80) >DroSim_CAF1 38846 108 + 1 GCAUCGGUGGCGUAUACGUGAUGCGGCUUAAUAAGGGAGCAUACUUGUCCCCUAACUGAAAUC------UGAAUGCCACAAUUGCAUUUCACACAGCCCACCAAAGUGUCACCA .....(((((((......((.((.((((.....((((..((....))..))))..........------.((((((.......)))))).....)))))).))...))))))). ( -29.80) >DroEre_CAF1 49385 108 + 1 GCAUCGGUGGCGUAUACGUGAUGUGGCUAAAUAAGGGAGCAUACUUGUCCACUAACCAACAUG------CGAAUGCGACAAUUGCAUUUCACACAGCCCACCAAAGUGUCACCA .....(((((((......((.((.((((.....(((((.((....))))).))........((------.((((((((...))))))))))...)))))).))...))))))). ( -27.30) >DroYak_CAF1 39218 114 + 1 GCAUCGGUGGCGUAUACGUGAUGUGGCUAAAUAAGGGAGCAUACUUGUCCACUAACUAAAAUCUGAAUCUGAAUGCCACAAAUGCAUUUCACGCAGUCCACCAAAGUGUCGCCA .....(((((((.....(((.((((((......(((((.((....))))).))........((.......))..))))))..(((.......)))...))).....))))))). ( -27.40) >consensus GCAUCGGUGGCGUAUACGUGAUGCGGCUUAAUAAGGGAGCAUACUUGUCCCCUAACUGAAAUC______UGAAUGCCACAAUUGCAUUUCACACAGCCCACCAAAGUGUCACCA .....(((((((......((.((.((((.....((((..((....))..)))).................((((((.......)))))).....)))))).))...))))))). (-25.88 = -26.16 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:20 2006