
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,544,802 – 7,544,964 |
| Length | 162 |
| Max. P | 0.924955 |

| Location | 7,544,802 – 7,544,896 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 95.77 |
| Mean single sequence MFE | -18.33 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.54 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.924955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
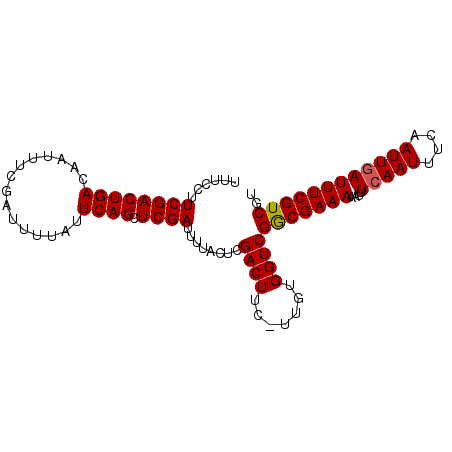
>X_DroMel_CAF1 7544802 94 + 22224390 UUUCCUUCGACUGACAAUUUCGAUUUUAUUCAGCUCGAUUUUACUCGACUUC-UUGUGGUCGGCGAAAAAUUUCAAUUUCAAUUGAUUUCGUCGU ......((((((((...............)))).))))........((((..-....))))(((((((....(((((....)))))))))))).. ( -19.06) >DroSec_CAF1 37890 94 + 1 UUUCCUUCGACUGACAAUUUCGAUUUUAUUCAGCUCGAUUUUACUCGACUUC-UUGUGGUCGGCGAAAAAUUUCAAUUUCAAUUAAUUUCGUCGU ......((((((((...............)))).))))........((((..-....))))(((((((((((........))))..))))))).. ( -16.06) >DroSim_CAF1 38272 94 + 1 UUUCCUUCGACUGACAAUUUCGAUUUUAUUCAGCUCGAUUUUACUCGACUUC-UUGUGGUCGGCGAAAAAUUUCAAUUUCAAUUGAUUUCGUCGU ......((((((((...............)))).))))........((((..-....))))(((((((....(((((....)))))))))))).. ( -19.06) >DroYak_CAF1 38612 92 + 1 UUU---UCGACUGACAAUUUCGAUUUUAUUCAGCUCGAUUUUACUCGACUUUUUUUUGGUCGACGAAAAAUUUCAAUUUCAAUUGAUUUCGUCGU ...---((((((((...............)))).))))........((((.......))))(((((((....(((((....)))))))))))).. ( -19.16) >consensus UUUCCUUCGACUGACAAUUUCGAUUUUAUUCAGCUCGAUUUUACUCGACUUC_UUGUGGUCGGCGAAAAAUUUCAAUUUCAAUUGAUUUCGUCGU ......((((((((...............)))).))))........((((.......))))(((((((....(((((....)))))))))))).. (-17.47 = -17.54 + 0.06)

| Location | 7,544,857 – 7,544,964 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
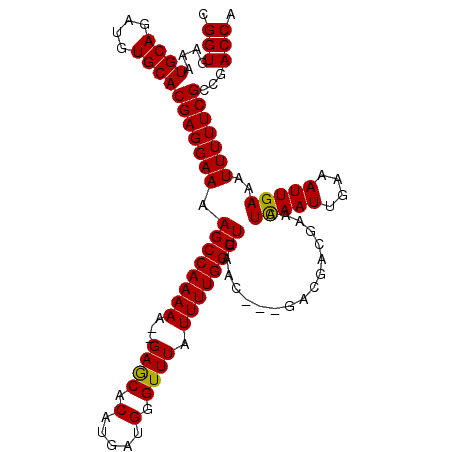
>X_DroMel_CAF1 7544857 107 - 22224390 CGGUGAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAA--GAGCACAUGAUGGGUUUAUUUUGGUUGAAC---GACGACGAAAUCAAUUGAAAUUGAAAUUUUUCGCCGACCA ((((((((..((.((((((.(..((.....)).....--).))))))..))..((.((((..(((((.(---(....))...)))))..)))).))....)))))))).... ( -26.00) >DroSec_CAF1 37945 107 - 1 CGGUGAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAA--GAGCACAUGAUGGGUUUAUUUUGGUUGAAC---GACGACGAAAUUAAUUGAAAUUGAAAUUUUUCGCCGACCA ((((((((((((....))))((......((((((((.--((((.(.....).)))).))))))))....---..))........................)))))))).... ( -24.50) >DroSim_CAF1 38327 107 - 1 CGGUGAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAA--GAGCACAUGAUGGGUUUAUUUUGGUUGAAC---GACGACGAAAUCAAUUGAAAUUGAAAUUUUUCGCCGACCA ((((((((..((.((((((.(..((.....)).....--).))))))..))..((.((((..(((((.(---(....))...)))))..)))).))....)))))))).... ( -26.00) >DroEre_CAF1 48854 109 - 1 CGGUAAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAGGGGAACACAUGAUGGGUUUAUUUUGGUUGAAC---GACGACGAAAUCAAUUGAAAUUGAAAUUUUUCGGCGACCA .(((....((((....))))(((((((.((((((((...((((.(.....).)))).))))))))....---..........(((((....)))))..)))))))...))). ( -23.70) >DroYak_CAF1 38665 110 - 1 CGGUGAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAA--GAACACAUGAUGGGUUUAUUUUGGUUGAGCGACGACGACGAAAUCAAUUGAAAUUGAAAUUUUUCGUCGACCA .(((....((((....))))((..(...((((((((.--((((.(.....).)))).))))))))...)..)).(((((((((((((....)))))....))))))))))). ( -29.50) >consensus CGGUGAAAUGCAGAUGUGCACGAGGAAAAGCCAAAAA__GAGCACAUGAUGGGUUUAUUUUGGUUGAAC___GACGACGAAAUCAAUUGAAAUUGAAAUUUUUCGCCGACCA .(((....((((....))))(((((((.((((((((...((((.(.....).)))).)))))))).................(((((....)))))..)))))))...))). (-21.78 = -21.38 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:18 2006