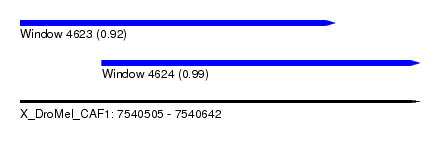
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,540,505 – 7,540,642 |
| Length | 137 |
| Max. P | 0.987955 |

| Location | 7,540,505 – 7,540,613 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.06 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -8.30 |
| Energy contribution | -8.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7540505 108 + 22224390 AAG----------AAGGGUGAAUGAAAAGUAA--GUUUGGAACUCUGCGCAAGUAUGUCGAUUUGUAUUAUAUUUUAUGCAAAUCCAGCUUAUUCUAUAAAUAUCCACAAAGAAUUAAAC ...----------..(((((.......(((((--(((.(((....(((....)))..))(((((((((........)))))))))))))))))).......))))).............. ( -21.74) >DroSec_CAF1 33629 91 + 1 AAG----------AAGGGUGCAUGAAAAGUAA------------------AAGU-UUUCGAUUUGUAUUAUAUUUUAUGCAAAUCCACCAUCUUCUAUAAAUAUUCACAAAGAAUAAAAC .((----------(((((((...(((((....------------------...)-))))(((((((((........)))))))))))))..))))).....(((((.....))))).... ( -20.50) >DroSim_CAF1 34004 91 + 1 AAG----------AAGGGUGCAUGAAAAGUAA------------------AAGU-UUUCGAUUUGUAUUAUAUUUUAUGCAAAUCCACCAUCUUCUAUAAAUAUUCACAAAGAAUAAAAC .((----------(((((((...(((((....------------------...)-))))(((((((((........)))))))))))))..))))).....(((((.....))))).... ( -20.50) >DroYak_CAF1 34373 109 + 1 AGGGUGCAUCAAAUAUAGUGCAUGAAAAGUAAAACUUUGAAACUCUGCGCAAGUAUGCCGAUUUGUAUUAUAUUUUAUGCAAAUCCACAA---UCUAUAAAUACC--------AUCCUAA .((((((((.((((((((((((...((((.....)))).......(((....)))........)))))))))))).)))))..)))....---............--------....... ( -23.80) >consensus AAG__________AAGGGUGCAUGAAAAGUAA__________________AAGU_UGUCGAUUUGUAUUAUAUUUUAUGCAAAUCCACCAUCUUCUAUAAAUAUCCACAAAGAAUAAAAC ...........................................................(((((((((........)))))))))................................... ( -8.30 = -8.30 + -0.00)



| Location | 7,540,533 – 7,540,642 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.34 |
| Mean single sequence MFE | -15.86 |
| Consensus MFE | -8.18 |
| Energy contribution | -8.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7540533 109 + 22224390 AACUCUGCGCAAGUAUGUCGAUUUGUAUUAUAUUUUAUGCAAAUCCAGCUUAUUCUAUAAAUAUCCACAAAGAAUUAAACAU----GUAUAUUUGGCAUAAACUAUUUACCAA .....(((.((((((((.((((((((((........)))))))))......(((((..............))))).......----))))))))))))............... ( -17.64) >DroSim_CAF1 34026 98 + 1 ----------AAGU-UUUCGAUUUGUAUUAUAUUUUAUGCAAAUCCACCAUCUUCUAUAAAUAUUCACAAAGAAUAAAACAU----GUAUGUCUGGCAUAAACUAUUUACCAA ----------..((-((..(((((((((........)))))))))................(((((.....)))))))))..----.((((.....))))............. ( -13.30) >DroEre_CAF1 44308 110 + 1 AACUCUGCGUAAGUAUGUCGAUUUGUAUUUUAUUUAAUGCAAAUCCAUCA---ACUAUAAAUAUCUACAAAGCAUAAAAUAUAAACAUAUAUUUGGCAUAAGCUAUUUACCAA .....((.(((((((.((.((((((((((......)))))))))).....---..................((...(((((((....))))))).))....))))))))))). ( -20.60) >DroYak_CAF1 34413 90 + 1 AACUCUGCGCAAGUAUGCCGAUUUGUAUUAUAUUUUAUGCAAAUCCACAA---UCUAUAAAUACC--------AUCCUAAAUACCCAUCUAAU-------ACCUAU-----AA .....(((....)))((..(((((((((........)))))))))..)).---............--------....................-------......-----.. ( -11.90) >consensus AACUCUGCGCAAGUAUGUCGAUUUGUAUUAUAUUUUAUGCAAAUCCACCA___UCUAUAAAUAUCCACAAAGAAUAAAACAU____AUAUAUUUGGCAUAAACUAUUUACCAA ...................(((((((((........))))))))).................................................................... ( -8.18 = -8.18 + -0.00)

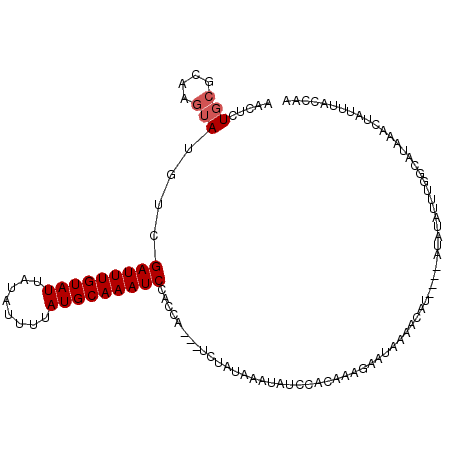
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:16 2006