
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,535,988 – 7,536,097 |
| Length | 109 |
| Max. P | 0.524948 |

| Location | 7,535,988 – 7,536,097 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -11.42 |
| Energy contribution | -10.81 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7535988 109 - 22224390 GCCAACCCUGUUGGCGUCAUAAAAAUGCCACAAAAAGCAAACACGCGUAAAUUAGAAGUAACAACAAAAA-AGUAUAAAAACACACAAAAAAAAUUUGUUCGAUUGCGUA ((((((...))))))..........(((........))).....((((((....................-.((......))..(((((.....)))))....)))))). ( -15.10) >DroSec_CAF1 29158 106 - 1 ACCAACCCUGUUGGCGUCAUAAAAAUGCCACAAAAAGCAAACACGCGUAAAUUAGAAGUAACAACAAAAACAGUAUAAAAACA--CA--AAAAAUGUCUUCGAUUGCGUA ........(((.(((((.......))))))))............((((((....((((..............((......))(--((--.....)))))))..)))))). ( -13.60) >DroSim_CAF1 29506 107 - 1 GCCAACCCUGUUGGCGUCAUAAAAAUGCCACAAAAAGCAAACACGCGUAAAUUAGAAGUAACAACAAAAA-AGUAUAAAAACACACA--AAAAAUGUCUUCGAUUGCGUA ((((((...))))))..........(((........))).....((((((....((((............-.((......)).....--........))))..)))))). ( -16.47) >DroEre_CAF1 39710 109 - 1 GCCAACCCCGUUGGCGUCAUAAAAAUGUCACAAAAAGCAAACACGCGUAAAUUAGAAUUAACAACAGAAA-AGAAUAAAAACACACACAAAAAAUGUAUUCGAUUGCGUA ((((((...))))))(.(((....))).).............((((((.(((....))).))........-.(((((...................)))))....)))). ( -14.21) >DroYak_CAF1 29872 97 - 1 GCCAACCCCGUUGGCGUCAUAAAAAUGCCACAAAAAGCAAACACGCGUAAAUUAGAAGUAACAACAAAAA-AGAAUAA------------AAAAUGUCUUCGAUUGCGUA ((((((...))))))..........(((........))).....((((((....((((..(((.......-.......------------....)))))))..)))))). ( -16.17) >DroAna_CAF1 36795 100 - 1 ---CACGCUUCUGGCGUCAUAGAAACGCCACAA-AGGCAAACACGCGUGAAUUGGAGGCGAAAAUGAAGG------AAAGGCAGAACGAAAAUGUGUCUUCGGUUGCGUA ---...((((.((((((.......))))))...-))))......((((.(((((((((((........(.------..........).......))))))))))))))). ( -25.16) >consensus GCCAACCCUGUUGGCGUCAUAAAAAUGCCACAAAAAGCAAACACGCGUAAAUUAGAAGUAACAACAAAAA_AGUAUAAAAACACACA__AAAAAUGUCUUCGAUUGCGUA ...........((((((.......))))))..............((((((....((((.......................................))))..)))))). (-11.42 = -10.81 + -0.61)

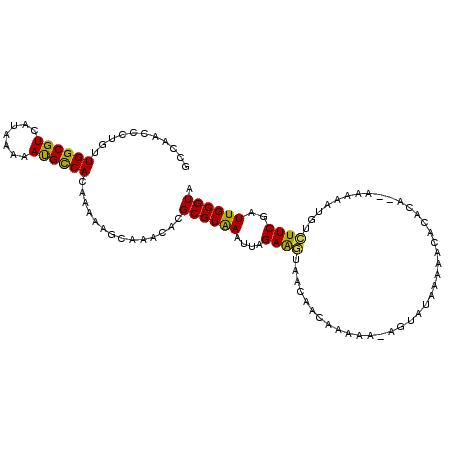
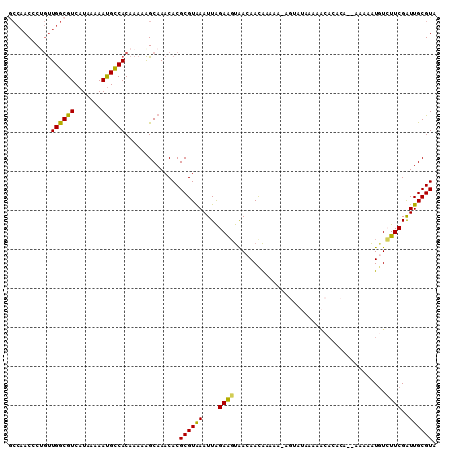
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:13 2006