
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,518,885 – 7,519,018 |
| Length | 133 |
| Max. P | 0.949951 |

| Location | 7,518,885 – 7,518,999 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -57.73 |
| Consensus MFE | -43.28 |
| Energy contribution | -43.47 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7518885 114 + 22224390 UUCCCUAUCCGCCGUACUUUGCUGCCGCUGCCAUUUUGGGUCGCAGUCUGGCGGGGAUUCCCGGUGCGGCAGCGGCCGCUGGAGCAGCUGCAGCA---GCCGCCGCAGUUGGCGCCU .........(((((.(((..((((((((.((((..(((.....)))..))))(((....)))...))))))))(((.((((..((....))..))---)).)))..))))))))... ( -53.00) >DroGri_CAF1 108225 117 + 1 UUCCCUACCCACCGUAUUUCGCUGCUGCUGCACUGCUCGGUCGCAGCCUGGCCGGCAUACCGGGUGCGGCGGCUGCUGCCGGUGCAGCGGCAGCUGCGGCUGCCGCAGUGGGUGCCG .....(((((((........(((((((((((((....((((.((((((..((((.(((.....))))))))))))).)))))))))))))))))(((((...))))))))))))... ( -70.00) >DroEre_CAF1 21314 114 + 1 UUCCCUAUCCGCCGUACUUUGCUGCCGCCGCCAUUUUGGGCCGCAGUCUGGCGGGGAUUCCCGGCGCAGCAGCGGCCGCUGGAGCAGCUGCAGCU---GCUGCCGCAGUUGGCGCCU .........(((((.(((.((((((.(((((((..(((.....)))..))))(((....))))))))))))(((((......((((((....)))---))))))))))))))))... ( -55.40) >DroYak_CAF1 15069 114 + 1 UACCCUAUCCGCCCUACUUUGCGGCCGCUGCCAUUUUGGGACGCAGUCUGGCGGGGAUUCCCGGUGCCGCUGCGGCCGCUGGAGCAGCUGCAGCA---GCUGCCGCUGUUGGCGCCA .........((((..((...(((((.(((((((..(((.....)))..))))(((....)))......((((((((.((....)).)))))))))---)).))))).)).))))... ( -52.50) >consensus UUCCCUAUCCGCCGUACUUUGCUGCCGCUGCCAUUUUGGGUCGCAGUCUGGCGGGGAUUCCCGGUGCGGCAGCGGCCGCUGGAGCAGCUGCAGCA___GCUGCCGCAGUUGGCGCCU .........(((((......(((((.(((((.......((((((.(((((.((((....)))).)).))).))))))......))))).(((((....))))).))))))))))... (-43.28 = -43.47 + 0.19)

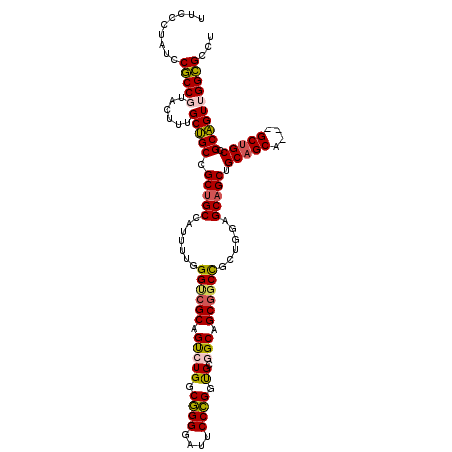
| Location | 7,518,885 – 7,518,999 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -56.83 |
| Consensus MFE | -43.74 |
| Energy contribution | -43.05 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7518885 114 - 22224390 AGGCGCCAACUGCGGCGGC---UGCUGCAGCUGCUCCAGCGGCCGCUGCCGCACCGGGAAUCCCCGCCAGACUGCGACCCAAAAUGGCAGCGGCAGCAAAGUACGGCGGAUAGGGAA ...((((...((((((.((---((..((....))..)))).)))((((((((...(((....)))((((...((.....))...)))).))))))))...))).))))......... ( -54.60) >DroGri_CAF1 108225 117 - 1 CGGCACCCACUGCGGCAGCCGCAGCUGCCGCUGCACCGGCAGCAGCCGCCGCACCCGGUAUGCCGGCCAGGCUGCGACCGAGCAGUGCAGCAGCAGCGAAAUACGGUGGGUAGGGAA ....(((((((((((...)))).(((((.((((((((((..((((((((((((.......)).))))..))))))..)))....))))))).))))).......)))))))...... ( -63.50) >DroEre_CAF1 21314 114 - 1 AGGCGCCAACUGCGGCAGC---AGCUGCAGCUGCUCCAGCGGCCGCUGCUGCGCCGGGAAUCCCCGCCAGACUGCGGCCCAAAAUGGCGGCGGCAGCAAAGUACGGCGGAUAGGGAA ...((((..((((((.(((---(((....)))))))).))))..((((((((((((((....)))(((.......))).......))).)))))))).......))))......... ( -55.70) >DroYak_CAF1 15069 114 - 1 UGGCGCCAACAGCGGCAGC---UGCUGCAGCUGCUCCAGCGGCCGCAGCGGCACCGGGAAUCCCCGCCAGACUGCGUCCCAAAAUGGCAGCGGCCGCAAAGUAGGGCGGAUAGGGUA (((((((......))).((---((((((.(((((....))))).))))))))...(((....)))))))..((((...((.....))..))))((((........))))........ ( -53.50) >consensus AGGCGCCAACUGCGGCAGC___AGCUGCAGCUGCUCCAGCGGCCGCUGCCGCACCGGGAAUCCCCGCCAGACUGCGACCCAAAAUGGCAGCGGCAGCAAAGUACGGCGGAUAGGGAA ...((((....((((((((....))))).(((((....))))))))((((((..((((....))))...(.((((...........))))).))))))......))))......... (-43.74 = -43.05 + -0.69)

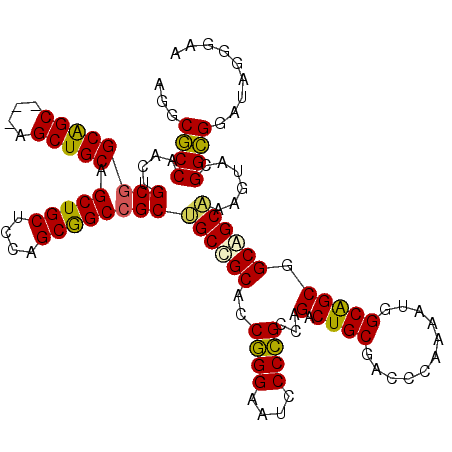
| Location | 7,518,925 – 7,519,018 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -35.72 |
| Energy contribution | -36.17 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7518925 93 - 22224390 UGCAACUCAUCACCACCAGAGGCGCCAACUGCGGCGGCUGCUGCAGCUGCUCCAGCGGCCGCUGCCGCACCGGGAAUCCCCGCCAGACUGCGA .(((..((.((.......))(((((.....(((((((((((((.((...)).))))))))))))).))...(((....)))))).)).))).. ( -39.50) >DroEre_CAF1 21354 93 - 1 UGCAACUCAUCGCCGCCGGAGGCGCCAACUGCGGCAGCAGCUGCAGCUGCUCCAGCGGCCGCUGCUGCGCCGGGAAUCCCCGCCAGACUGCGG .(((......((((......))))....(((((((.(((((.((.(((((....))))).)).))))))))(((....)))).)))..))).. ( -44.00) >DroYak_CAF1 15109 93 - 1 UGCAACUCAUCACCGCCAGUGGCGCCAACAGCGGCAGCUGCUGCAGCUGCUCCAGCGGCCGCAGCGGCACCGGGAAUCCCCGCCAGACUGCGU .............(((.(((((((((......))).((((((((.(((((....))))).))))))))...(((....))))))..)))))). ( -44.80) >consensus UGCAACUCAUCACCGCCAGAGGCGCCAACUGCGGCAGCUGCUGCAGCUGCUCCAGCGGCCGCUGCCGCACCGGGAAUCCCCGCCAGACUGCGA .(((..((.....((((...))))........(((.(((((.((.(((((....))))).)).)))))...(((....)))))).)).))).. (-35.72 = -36.17 + 0.45)


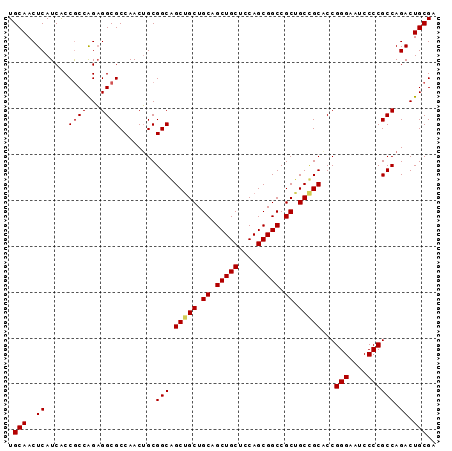
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:09 2006