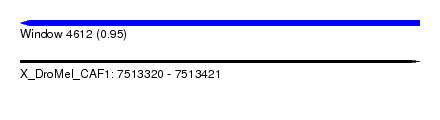
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,513,320 – 7,513,421 |
| Length | 101 |
| Max. P | 0.949874 |

| Location | 7,513,320 – 7,513,421 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.37 |
| Mean single sequence MFE | -17.08 |
| Consensus MFE | -14.60 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7513320 101 - 22224390 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUUUUUCCUUUUCCCCCU-------ACUCAG----UGACUCCUUUG ..(((((.......((((((......)))))).((((.....))))........)))))((((((.((................-------))..))----))))....... ( -17.09) >DroPse_CAF1 62791 104 - 1 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUAUUUCUUUUCUCCCC--------AUUCGAAUGAGGACUCCUCCA .((((((.......((((((......)))))).((((.....))))........))))))(((((((................--------...)))))))(((....))). ( -20.21) >DroGri_CAF1 101200 80 - 1 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUUGUUCAAUGUGU-------------------------------- (((((((.......((((((......)))))).((((.....))))........)))))))...................-------------------------------- ( -14.60) >DroWil_CAF1 24170 76 - 1 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUUUUUUUCU------------------------------------ (((((((.......((((((......)))))).((((.....))))........)))))))...............------------------------------------ ( -14.60) >DroYak_CAF1 8974 108 - 1 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUUUUUCCUUUUCCCUUUUCCCCCAACUCAC----UGGCUCCUUUG (((((((.......((((((......)))))).((((.....))))........)))))))...........................(((......----)))........ ( -15.80) >DroPer_CAF1 68118 104 - 1 AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUAUUUCUUUUCUCCCC--------AUUCGAAUGAGGACUCCUCCA .((((((.......((((((......)))))).((((.....))))........))))))(((((((................--------...)))))))(((....))). ( -20.21) >consensus AUUGGCACACGCACGCAAUUUGCAUUAAUUGCAAGAAAAAUUUUCUAAUUAAAAUGCCAGUCAUUUGUUUUUCUUUUUCCCCC________A_UC_______GACUCCU___ (((((((.......((((((......)))))).((((.....))))........)))))))................................................... (-14.60 = -14.60 + -0.00)

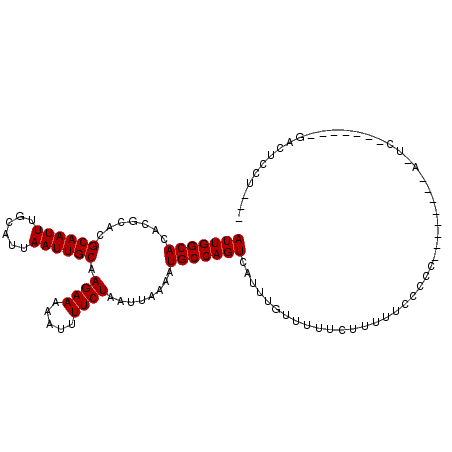

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:05 2006