
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,490,020 – 7,490,207 |
| Length | 187 |
| Max. P | 0.977480 |

| Location | 7,490,020 – 7,490,140 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.82 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7490020 120 - 22224390 UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGGUGGGCUAGUGCUGCUGCUGCUGCUGC (((((((......)))))))..(((((((..(((((......)))))..)))))))(((((.((((((..((((....))))...(((((((....)))))))))..)))).)).))).. ( -37.20) >DroSec_CAF1 23047 117 - 1 UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGGUGAGCUAGUGCUGCUG---CUGCUGC (((((((......)))))))..(((((((..(((((......)))))..)))))))(((((.((((.(((.........)))...((((((......)))))).)))).)---).))).. ( -33.90) >DroSim_CAF1 25039 117 - 1 UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGGUGGGCUAGUGCUGCUG---CUGCUGC (((((((......)))))))..(((((((..(((((......)))))..)))))))(((((.((((.(((.........)))...(((((((....))))))).)))).)---).))).. ( -36.60) >DroEre_CAF1 24147 117 - 1 UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAACCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGGUGGGCUAGUGCUGCUG---CUUCUGC (((((((......))))))).....(((((((.(((((....((...((....)).))..)))))((((.((((....))))...(((((((....)))))))..)))))---)))))). ( -32.10) >DroYak_CAF1 26864 106 - 1 UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAACCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGUUGGGCUAGUGCU-------------- (((((((......)))))))..........((((((((....((...((....)).))..)))))))).................(((((((....)))))))...-------------- ( -27.10) >consensus UGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUUUGGAAAUUAGUCGGUGGGCUAGUGCUGCUG___CUGCUGC (((((((......)))))))..(((((((..(((((......)))))..)))))))(((((((....))))..............(((((((....)))))))))).............. (-28.38 = -28.98 + 0.60)

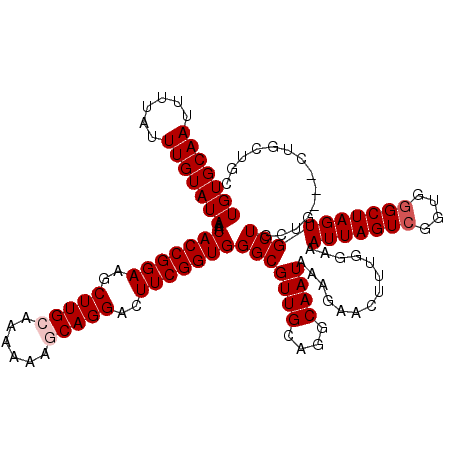
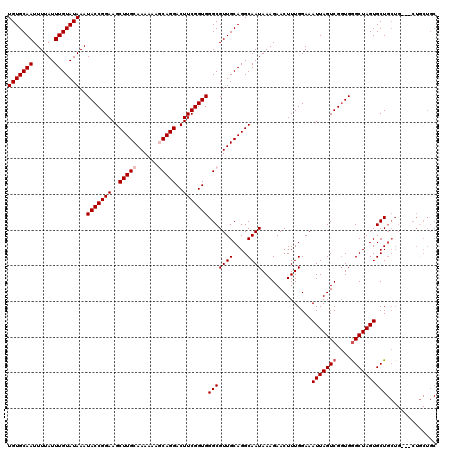
| Location | 7,490,060 – 7,490,173 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.03 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -28.42 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7490060 113 - 22224390 UCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU ...........((((-----.......(((.((...--(((((((((......)))))))))..)).)))((((((((....((...((....))..)).))))))))......)))).. ( -33.80) >DroSec_CAF1 23084 113 - 1 UCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU ...........((((-----.......(((.((...--(((((((((......)))))))))..)).)))((((((((....((...((....))..)).))))))))......)))).. ( -33.80) >DroSim_CAF1 25076 113 - 1 UCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU ...........((((-----.......(((.((...--(((((((((......)))))))))..)).)))((((((((....((...((....))..)).))))))))......)))).. ( -33.80) >DroEre_CAF1 24184 113 - 1 UCCGUUGAUUUGUUC-----GGUUUGGCCUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAACCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU ...........((((-----((.....))...(((.--(((((((((......))))))))).)))....((((((((....((...((....)).))..))))))))......)))).. ( -33.40) >DroYak_CAF1 26890 120 - 1 UCCGUUGAUUUGCUCAGUUUGGUUUGGUUUGGGGUUUUUUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAACCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU (((........((.(((......)))))....(((...(((((((((......))))))))).)))))).((((((((....((...((....)).))..))))))))............ ( -30.80) >consensus UCCGUUGAUUUGUUC_____GGUUUGGUUUGGGGUU__UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAAAAGCAGGACUUCGGUGGGCGUUGCAGGCAAUAAAGAACUU ...(((..((((((..................(((...(((((((((......))))))))).)))....((((((((....((...((....))..)).)))))))))))))).))).. (-28.42 = -29.02 + 0.60)

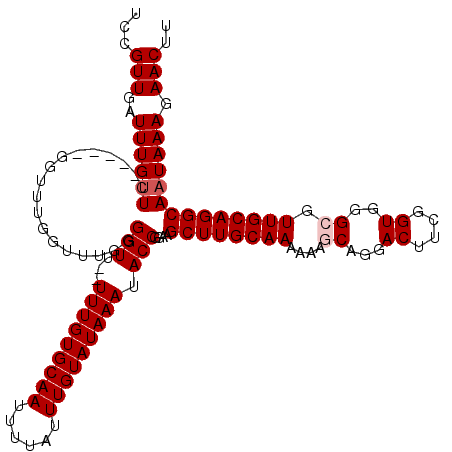

| Location | 7,490,100 – 7,490,207 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.77 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -24.30 |
| Energy contribution | -24.90 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7490100 107 - 22224390 ------AAGCACCAAAUGGCAAUGACUUCUGCUUCGAUUUUCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA ------((((.((((((((((........)))).(((....(((..........)-----)).)))))))))(((.--(((((((((......))))))))).)))....))))...... ( -26.90) >DroSec_CAF1 23124 107 - 1 ------AAGCACCAAAUGGCAAUGACUUUUGCUUCGAUUUUCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA ------((((.(((((((((((......))))).(((....(((..........)-----)).)))))))))(((.--(((((((((......))))))))).)))....))))...... ( -28.60) >DroSim_CAF1 25116 107 - 1 ------AAGCACCAAAUGGCAAUGACUUUUGCUUCGAUUUUCCGUUGAUUUGUUC-----GGUUUGGUUUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA ------((((.(((((((((((......))))).(((....(((..........)-----)).)))))))))(((.--(((((((((......))))))))).)))....))))...... ( -28.60) >DroEre_CAF1 24224 113 - 1 ACACCAAAACACCAAAUGGCAAUGACUUUUGCUUCGAUUUUCCGUUGAUUUGUUC-----GGUUUGGCCUGGGGUU--UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA ...((......(((...(((...((((...((.(((((.....)))))...))..-----))))..))))))(((.--(((((((((......))))))))).)))))............ ( -27.50) >DroYak_CAF1 26930 118 - 1 G--CCCAAGCACCAAAUGGCAAUGACUUUUGCUUCGAUUUUCCGUUGAUUUGCUCAGUUUGGUUUGGUUUGGGGUUUUUUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA .--..(((((.(((((((((...((((...((.(((((.....)))))...))..))))..)))..))))))(((...(((((((((......))))))))).)))....)))))..... ( -31.50) >consensus ______AAGCACCAAAUGGCAAUGACUUUUGCUUCGAUUUUCCGUUGAUUUGUUC_____GGUUUGGUUUGGGGUU__UUUGUGCAAUUUUAUUUGUAUAAAUACCGGAAGCUUGCAAAA ......((((((((((((((((......)))))(((((.....))))).............)))))))....(((...(((((((((......))))))))).)))....))))...... (-24.30 = -24.90 + 0.60)

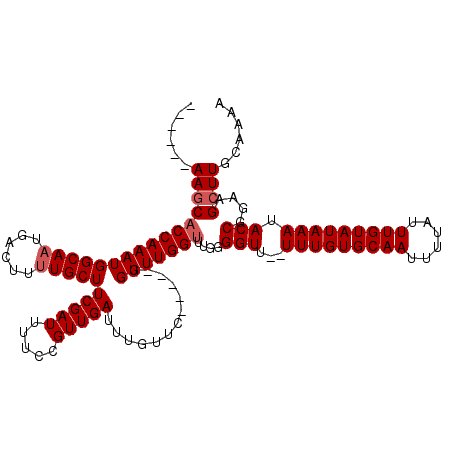
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:58 2006