
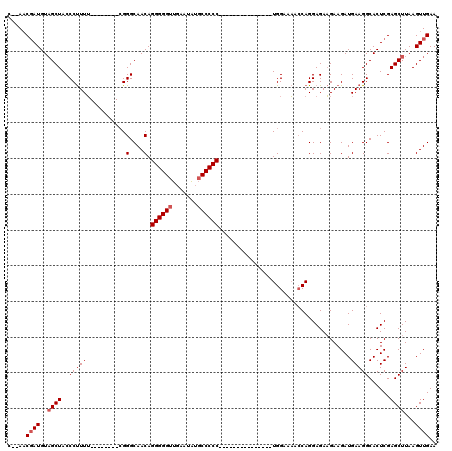
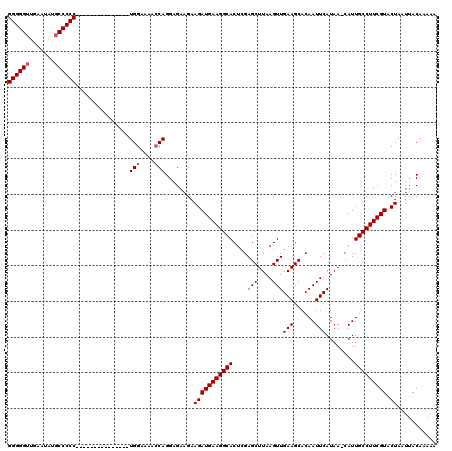
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,487,782 – 7,488,008 |
| Length | 226 |
| Max. P | 0.992723 |

| Location | 7,487,782 – 7,487,877 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -19.32 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487782 95 + 22224390 C--AACGAUGUAGCUACCCUUUU--------CGGGCAACAGGGGGUUGAAUAUGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAA (--(((.....((((...(((((--------(.((...((((((((.......))))))---------------)).....))..))))))....(((......)))))))...)))).. ( -31.00) >DroSec_CAF1 20810 94 + 1 C--AACGAUGUAGCUACCCUUUU--------CGGGCAACAGGGGGUUGAAUAU-CCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAA (--(((.....((((...(((((--------(.((...(((((((........-)))))---------------)).....))..))))))....(((......)))))))...)))).. ( -29.50) >DroSim_CAF1 22807 95 + 1 C--AACGAUGUAGCUACCCUUUU--------CGGGCAACAGGGGGUUGAAUAUGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAA (--(((.....((((...(((((--------(.((...((((((((.......))))))---------------)).....))..))))))....(((......)))))))...)))).. ( -31.00) >DroEre_CAF1 21831 95 + 1 C--AACCAUGUAGCUACCCUUUU--------CGGGCAACAGGGGGUUGAACAGGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAA (--(((.....((((...(((((--------(.((...(((((((((.....)))))))---------------)).....))..))))))....(((......)))))))...)))).. ( -30.70) >DroYak_CAF1 24369 120 + 1 CCUACCGAUGUUGCUACCCCUUGGCGGCAACAGGGCAACAGGGGGUUGAAUGUGCCCCCUUUCUUUUUUUUUUUUGGAAUAGCAGGAGAUGAAGAUGAAGGCACUCGAGCUUAAGUUGAA ....((..((((((..((....))..)))))).))((((..(((.((((..(((((.....(((((...(((((((......))))))).)))))....))))))))).)))..)))).. ( -38.80) >consensus C__AACGAUGUAGCUACCCUUUU________CGGGCAACAGGGGGUUGAAUAUGCCCCC_______________UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAA .....((((..((((..(((((...........(....).((((((.......)))))).....................................)))))......))))...)))).. (-19.32 = -20.12 + 0.80)


| Location | 7,487,782 – 7,487,877 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.79 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487782 95 - 22224390 UUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCAUAUUCAACCCCCUGUUGCCCG--------AAAAGGGUAGCUACAUCGUU--G ..((((........((((((((............(((((....)))---------------))))))))..))........(((((((.--------....)))))))......)))--) ( -27.20) >DroSec_CAF1 20810 94 - 1 UUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGG-AUAUUCAACCCCCUGUUGCCCG--------AAAAGGGUAGCUACAUCGUU--G ..((((........(((((...........(((((((((....)))---------------)))))-))))))........(((((((.--------....)))))))......)))--) ( -27.70) >DroSim_CAF1 22807 95 - 1 UUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCAUAUUCAACCCCCUGUUGCCCG--------AAAAGGGUAGCUACAUCGUU--G ..((((........((((((((............(((((....)))---------------))))))))..))........(((((((.--------....)))))))......)))--) ( -27.20) >DroEre_CAF1 21831 95 - 1 UUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCCUGUUCAACCCCCUGUUGCCCG--------AAAAGGGUAGCUACAUGGUU--G ....((((......)))).............((((((((....)))---------------)))))......(((((....(((((((.--------....))))))).....))))--) ( -29.20) >DroYak_CAF1 24369 120 - 1 UUCAACUUAAGCUCGAGUGCCUUCAUCUUCAUCUCCUGCUAUUCCAAAAAAAAAAAAGAAAGGGGGCACAUUCAACCCCCUGUUGCCCUGUUGCCGCCAAGGGGUAGCAACAUCGGUAGG .........(((..(((...............)))..)))...((...............((((((.........))))))..((((.((((((..((....))..))))))..)))))) ( -31.96) >consensus UUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA_______________GGGGGCAUAUUCAACCCCCUGUUGCCCG________AAAAGGGUAGCUACAUCGUU__G ..............(((.(((................))).))).................(((((.........))))).(((((((.............)))))))............ (-20.59 = -20.79 + 0.20)

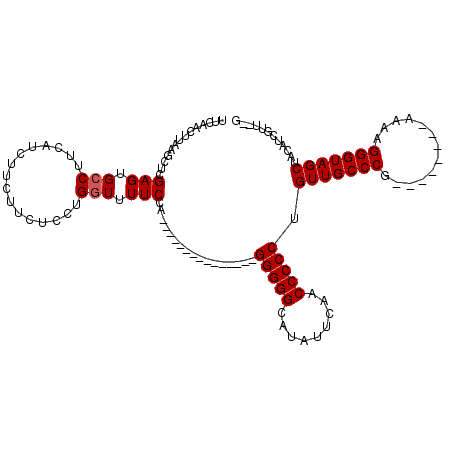

| Location | 7,487,812 – 7,487,916 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487812 104 + 22224390 GGGGGUUGAAUAUGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAA ((((((.......))))))---------------(((....)))........(((((((((((....(((....)))(((......)))....-...))))))))).))........... ( -27.40) >DroSec_CAF1 20840 104 + 1 GGGGGUUGAAUAU-CCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAACCAUUGCCUUCGUACUAAUGACAAAAA ((((((.....))-))))(---------------(((....)))).......(((((((((((....(((....)))(((......)))........))))))))).))........... ( -27.70) >DroSim_CAF1 22837 104 + 1 GGGGGUUGAAUAUGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAA ((((((.......))))))---------------(((....)))........(((((((((((....(((....)))(((......)))....-...))))))))).))........... ( -27.40) >DroEre_CAF1 21861 104 + 1 GGGGGUUGAACAGGCCCCC---------------UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAA (((((((.....)))))))---------------(((....)))........(((((((((((....(((....)))(((......)))....-...))))))))).))........... ( -28.20) >DroYak_CAF1 24409 119 + 1 GGGGGUUGAAUGUGCCCCCUUUCUUUUUUUUUUUUGGAAUAGCAGGAGAUGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAA ((((((.......))))))...((((...(((((((......))))))).))))(((((((((....(((....)))(((......)))....-...))))))))).............. ( -30.70) >consensus GGGGGUUGAAUAUGCCCCC_______________UGGAAAACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA_CAUUGCCUUCGUACUAAUUACAAAAA ((((((.......))))))...............(((....)))........(((((((((((....(((....)))(((......)))........))))))))).))........... (-24.40 = -24.80 + 0.40)

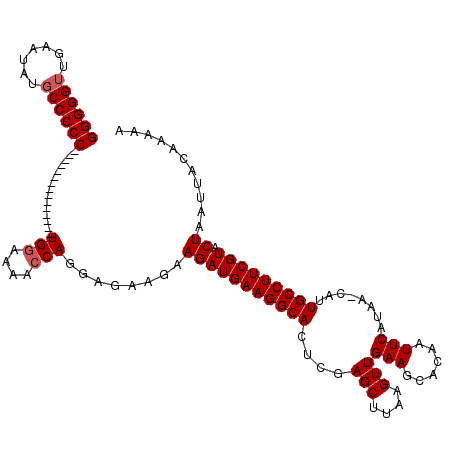
| Location | 7,487,812 – 7,487,916 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487812 104 - 22224390 UUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCAUAUUCAACCCCC ....(((.........(((((((...-.......(((.((((......)))).))).)))))))........(((((((....)))---------------)))))))............ ( -28.00) >DroSec_CAF1 20840 104 - 1 UUUUUGUCAUUAGUACGAAGGCAAUGGUUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGG-AUAUUCAACCCCC ....((((........(((((((...........(((.((((......)))).))).)))))))........(((((((....)))---------------)))))-))).......... ( -29.20) >DroSim_CAF1 22837 104 - 1 UUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCAUAUUCAACCCCC ....(((.........(((((((...-.......(((.((((......)))).))).)))))))........(((((((....)))---------------)))))))............ ( -28.00) >DroEre_CAF1 21861 104 - 1 UUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA---------------GGGGGCCUGUUCAACCCCC ...(((.(((..((..(((((((...-.......(((.((((......)))).))).)))))))........(((((((....)))---------------))))))..))))))..... ( -28.60) >DroYak_CAF1 24409 119 - 1 UUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCAUCUCCUGCUAUUCCAAAAAAAAAAAAGAAAGGGGGCACAUUCAACCCCC ((((((....(((((.(((((((...-.......(((.((((......)))).))).)))))))............)))))...))))))...........(((((.........))))) ( -25.22) >consensus UUUUUGUAAUUAGUACGAAGGCAAUG_UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGUUUUCCA_______________GGGGGCAUAUUCAACCCCC ................(((((((...........(((.((((......)))).))).))))))).....................................(((((.........))))) (-23.02 = -23.02 + -0.00)

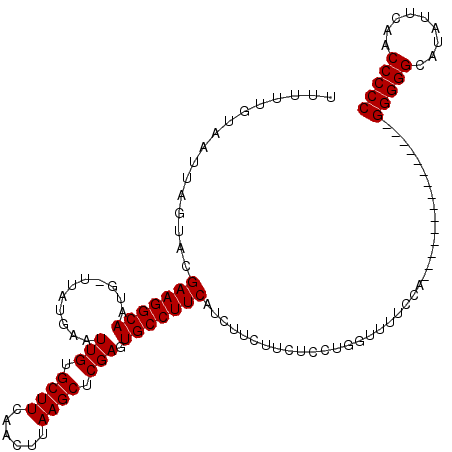

| Location | 7,487,837 – 7,487,953 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -23.64 |
| Consensus MFE | -20.90 |
| Energy contribution | -20.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487837 116 + 22224390 ACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAAGUGCUCUCGCAC-UCCUCCCUUUAAAAAAAAAAAAAGA-- ....((((....(((((((((((....(((....)))(((......)))....-...))))))))).))..........(((((....))))-).))))...................-- ( -25.40) >DroSec_CAF1 20864 115 + 1 ACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAACCAUUGCCUUCGUACUAAUGACAAAAAGUGCUCUCGCCCAACCUCCCUCCUAAAA-A--AAAAGA-- ...(((((........(((((((....(((....)))(((......)))........)))))))(((((..........)))))...............)))))....-.--......-- ( -22.50) >DroSim_CAF1 22862 116 + 1 ACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAAGUGCUCUCGCUGAACCUCCCUCCUAAAA-AAAAAAAGA-- ..(((((((.......(((((((....(((....)))(((......)))....-...)))))))(((((..........))))))))).)))................-.........-- ( -23.10) >DroEre_CAF1 21886 114 + 1 ACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAAGUGCUCUCGCCCAACCUCCCUUCUAAAA-A--AAAAGA-- ....((((........(((((((....(((....)))(((......)))....-...)))))))(((((..........)))))...........)))).........-.--......-- ( -20.90) >DroYak_CAF1 24449 116 + 1 AGCAGGAGAUGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA-CAUUGCCUUCGUACUAAUUACAAAAAGUGCUCUCGCCCAACCUCCCUGCUACAA-A--AAAAAAUU ((((((.((.(.....(((((((....(((....)))(((......)))....-...)))))))(((((..........)))))..........).))))))))....-.--........ ( -26.30) >consensus ACCAGGAGAAGAAGAUGAAGGCACUCGAGCUUAAGUUGAAGCACAAUUCAUAA_CAUUGCCUUCGUACUAAUUACAAAAAGUGCUCUCGCCCAACCUCCCUCCUAAAA_A__AAAAGA__ ....((((........(((((((....(((....)))(((......)))........)))))))(((((..........)))))...........))))..................... (-20.90 = -20.90 + 0.00)


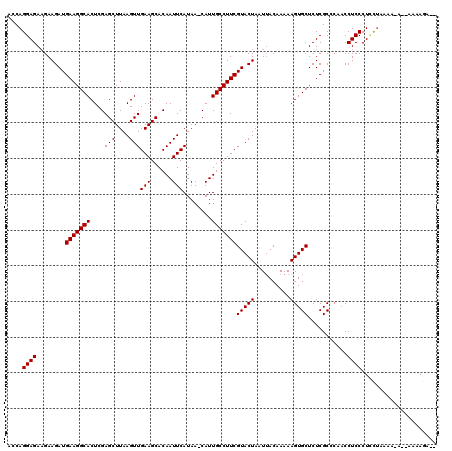
| Location | 7,487,837 – 7,487,953 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487837 116 - 22224390 --UCUUUUUUUUUUUUUAAAGGGAGGA-GUGCGAGAGCACUUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGU --..................(((((((-((((....))))))..............(((((((...-.......(((.((((......)))).))).)))))))........)))))... ( -32.30) >DroSec_CAF1 20864 115 - 1 --UCUUUU--U-UUUUAGGAGGGAGGUUGGGCGAGAGCACUUUUUGUCAUUAGUACGAAGGCAAUGGUUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGU --......--.-..((((((((((((..(((((.(((((((..........)))..(((((((((........))))).)))).......))))...)))))....)))).)))))))). ( -35.10) >DroSim_CAF1 22862 116 - 1 --UCUUUUUUU-UUUUAGGAGGGAGGUUCAGCGAGAGCACUUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGU --.........-..((((((((((((....((....))..................(((((((...-.......(((.((((......)))).))).)))))))..)))).)))))))). ( -32.00) >DroEre_CAF1 21886 114 - 1 --UCUUUU--U-UUUUAGAAGGGAGGUUGGGCGAGAGCACUUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGU --......--.-.....((..(((((..(((((.((((.......(((.....)))(((((((((.-......))))).)))).......))))...)))))....)))))..))..... ( -29.80) >DroYak_CAF1 24449 116 - 1 AAUUUUUU--U-UUGUAGCAGGGAGGUUGGGCGAGAGCACUUUUUGUAAUUAGUACGAAGGCAAUG-UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCAUCUCCUGCU ........--.-....((((((((((..(((((.((((.......(((.....)))(((((((((.-......))))).)))).......))))...)))))....))))....)))))) ( -33.20) >consensus __UCUUUU__U_UUUUAGAAGGGAGGUUGGGCGAGAGCACUUUUUGUAAUUAGUACGAAGGCAAUG_UUAUGAAUUGUGCUUCAACUUAAGCUCGAGUGCCUUCAUCUUCUUCUCCUGGU ....................((((((....((....))..................(((((((...........(((.((((......)))).))).))))))).......))))))... (-26.70 = -26.70 + -0.00)

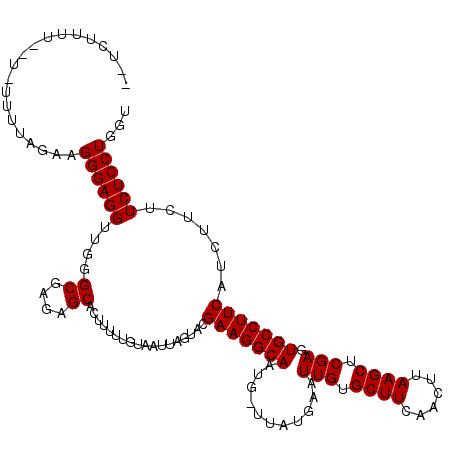
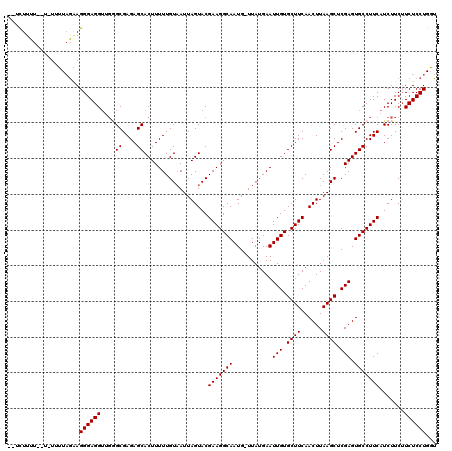
| Location | 7,487,916 – 7,488,008 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -19.16 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7487916 92 - 22224390 CCUCAGGAUUUCUGAGCUUCUGAAUUUCUGGAUUUCACGAUUCCCGCACUUUUCUUCUUUUUUUUUUUUUAAAGGGAGGA-GUGCGAGAGCAC .(((.(((.((.((((.(((.(.....).))).)))).)).)))(((((((((((.((((..........))))))))))-))))).)))... ( -26.30) >DroSec_CAF1 20944 90 - 1 CCUCAGGAUUUCUGAGCUUCUGAAUUUCUGUAUUUCACGAUUCCCGCACUUUUCUUCUUUU--U-UUUUAGGAGGGAGGUUGGGCGAGAGCAC .(((((.....)))))..((((((.........)))).))...(((.((((..(((((...--.-....)))))..)))))))((....)).. ( -24.20) >DroSim_CAF1 22941 92 - 1 CCUCAGGAUUUCUGAGCUUCUGAAUUUCUGGAUUUCACGAUUCCCGCACUUUUCUUCUUUUUUU-UUUUAGGAGGGAGGUUCAGCGAGAGCAC .(((.(((.((.((((.(((.(.....).))).)))).)).)))(((((((..(((((......-....)))))..))))...))).)))... ( -23.10) >DroEre_CAF1 21965 89 - 1 CCUCAGGAUUUCUGAGCU-CUGAAUUUCUGGGUUUCAUGAUUCCCGCACUUUUCUUCUUUU--U-UUUUAGAAGGGAGGUUGGGCGAGAGCAC .(((((((.(((......-..))).)))))))...........(((.((((..(((((...--.-....)))))..)))))))((....)).. ( -28.60) >consensus CCUCAGGAUUUCUGAGCUUCUGAAUUUCUGGAUUUCACGAUUCCCGCACUUUUCUUCUUUU__U_UUUUAGAAGGGAGGUUGGGCGAGAGCAC .(((.(((.((.((((.(((.(.....).))).)))).)).)))(((((((..(((((...........)))))..))))...))).)))... (-19.16 = -19.98 + 0.81)

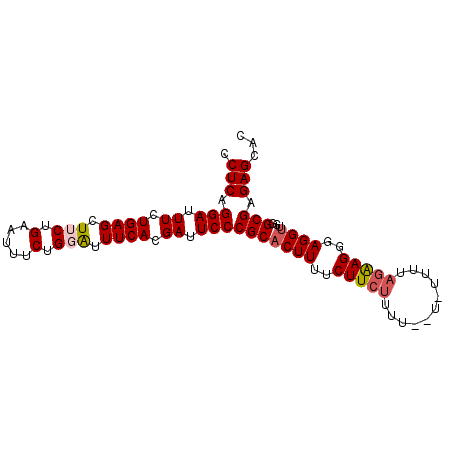
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:54 2006