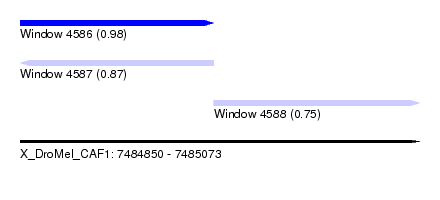
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,484,850 – 7,485,073 |
| Length | 223 |
| Max. P | 0.978583 |

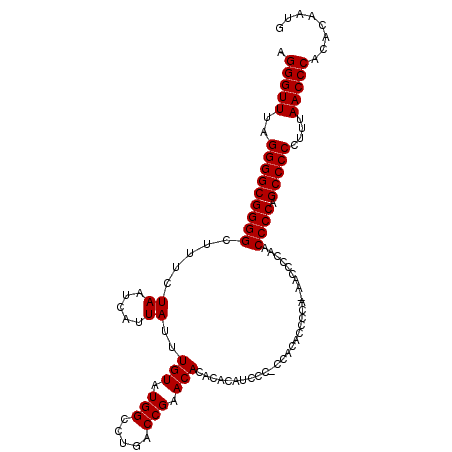
| Location | 7,484,850 – 7,484,958 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 97.54 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7484850 108 + 22224390 AGGGUUUAGGGGCGGGGCU-UCUAAUCAUUAUUUGUAUGGCCUGACCGAACACACACAUCCCCCCGCACCCCAAAACCCCAACCCCAGCCCCCUUUAACCCACACAAUG .(((((..(((((((((..-.............(((.(((.....))).))).............(.....)..........)))).)))))....)))))........ ( -26.80) >DroSec_CAF1 17918 107 + 1 AGGGUUUAGGGGCGGGGCUUUCUAAUCAUUAUUUGUAUGGCCUGACCGAACACACACAUCCC-CCACACCCCA-AACCCCAACCCCAGCCCCCUUUAACCCACACAAUG .(((((..(((((((((.....((.....))..(((.(((.....))).)))..........-..........-........)))).)))))....)))))........ ( -25.90) >DroSim_CAF1 19603 107 + 1 AGGGUUUAGGGGCGGGGCUUUCUAAUCAUUAUUUGUAUGGCCUGACCGAACACACACAUCCC-CCACACCCCA-AACCCCAACCCCAGCCCCCUUUAACCCACACAAUG .(((((..(((((((((.....((.....))..(((.(((.....))).)))..........-..........-........)))).)))))....)))))........ ( -25.90) >consensus AGGGUUUAGGGGCGGGGCUUUCUAAUCAUUAUUUGUAUGGCCUGACCGAACACACACAUCCC_CCACACCCCA_AACCCCAACCCCAGCCCCCUUUAACCCACACAAUG .(((((..(((((((((.....((.....))..(((.(((.....))).)))..............................)))).)))))....)))))........ (-25.90 = -25.90 + 0.00)

| Location | 7,484,850 – 7,484,958 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 97.54 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -37.67 |
| Energy contribution | -38.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7484850 108 - 22224390 CAUUGUGUGGGUUAAAGGGGGCUGGGGUUGGGGUUUUGGGGUGCGGGGGGAUGUGUGUGUUCGGUCAGGCCAUACAAAUAAUGAUUAGA-AGCCCCGCCCCUAAACCCU ................((((..(((((.(((((((((..(((.((..(.....((((((..(.....)..))))))..)..))))).))-))))))).)))))..)))) ( -37.20) >DroSec_CAF1 17918 107 - 1 CAUUGUGUGGGUUAAAGGGGGCUGGGGUUGGGGUU-UGGGGUGUGG-GGGAUGUGUGUGUUCGGUCAGGCCAUACAAAUAAUGAUUAGAAAGCCCCGCCCCUAAACCCU ........(((((...((((((.((((((...(((-((.(((.(((-.((((......))))..))).)))...)))))...........)))))))))))).))))). ( -39.84) >DroSim_CAF1 19603 107 - 1 CAUUGUGUGGGUUAAAGGGGGCUGGGGUUGGGGUU-UGGGGUGUGG-GGGAUGUGUGUGUUCGGUCAGGCCAUACAAAUAAUGAUUAGAAAGCCCCGCCCCUAAACCCU ........(((((...((((((.((((((...(((-((.(((.(((-.((((......))))..))).)))...)))))...........)))))))))))).))))). ( -39.84) >consensus CAUUGUGUGGGUUAAAGGGGGCUGGGGUUGGGGUU_UGGGGUGUGG_GGGAUGUGUGUGUUCGGUCAGGCCAUACAAAUAAUGAUUAGAAAGCCCCGCCCCUAAACCCU ........(((((...((((((.((((((..((((.....((((((...(((.((......)))))...)))))).......))))....)))))))))))).))))). (-37.67 = -38.00 + 0.33)


| Location | 7,484,958 – 7,485,073 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -19.84 |
| Energy contribution | -19.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7484958 115 + 22224390 CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCCGUUUUCUGCCUUGACUUGACCAUUCC-----CACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((.........)))).))).((((((....-----((((..........)))).....)))))))))))..)))))........ ( -21.80) >DroSec_CAF1 18025 115 + 1 CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCCGUUUUCUGCCUUGACUUGACCAUUCC-----CACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((.........)))).))).((((((....-----((((..........)))).....)))))))))))..)))))........ ( -21.80) >DroSim_CAF1 19710 115 + 1 CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCCGUUUUCUGCCUUGACUUGACCAUUCC-----CACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((.........)))).))).((((((....-----((((..........)))).....)))))))))))..)))))........ ( -21.80) >DroEre_CAF1 19041 115 + 1 CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCCGUUUUCUGCCUUGACUUGACCAUUUC-----CACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((.........)))).))).((((((....-----((((..........)))).....)))))))))))..)))))........ ( -21.80) >DroYak_CAF1 21492 120 + 1 CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCUGUUUUCUGCCUUGACUUGACCAUUUCCCCCCCACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((..(....).)))).))).((((((.........((((..........)))).....)))))))))))..)))))........ ( -20.04) >consensus CCCACAUAUGAGUUAUAUUUAUUUGUAUUUGUCUGGGCCCGUUUUCUGCCUUGACUUGACCAUUCC_____CACACCCUUUGCUUUGUGCUAUUUGGUCAGUGUAAUGCUUAAUUAUUUA ........((((((((((............(((.((((.........)))).))).((((((.........((((..........)))).....)))))))))))..)))))........ (-19.84 = -19.84 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:43 2006