
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,484,525 – 7,484,676 |
| Length | 151 |
| Max. P | 0.925653 |

| Location | 7,484,525 – 7,484,642 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.26 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
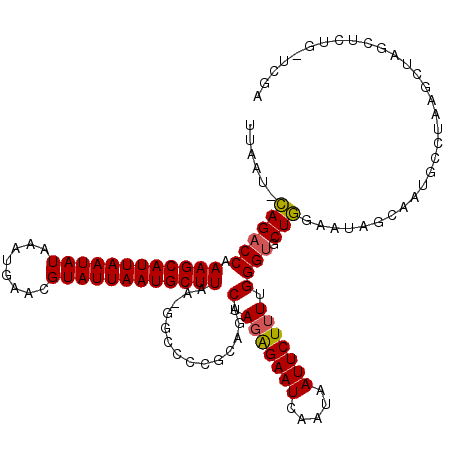
>X_DroMel_CAF1 7484525 117 - 22224390 UUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGGGAAUCAAUAAUUCUUUUGGGUGCUGAAAUAGCAAUGCCUAAGCUAGCUCUG-UCGA .....-....((.((((((((((((........))))))))))))..-))...(((((((((((((((.....)))))(((((((..((......))..))))))))))).))))-.)). ( -31.60) >DroSec_CAF1 17593 118 - 1 UUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUUUGGGUGCUGGAAUAGCAAUGCCUAAGCUAGCUCCGUUCGA ....(-(..((..((((((((((((........))))))))))))..-(((((((......(((((((.....))))))))))).)))(((.((((.........))))..)))))..)) ( -30.90) >DroSim_CAF1 19278 118 - 1 UUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUUUGGGUGCUGGAAUAGCAAUGCCUAAGCUAGCUCCGUUCGA ....(-(..((..((((((((((((........))))))))))))..-(((((((......(((((((.....))))))))))).)))(((.((((.........))))..)))))..)) ( -30.90) >DroEre_CAF1 18602 114 - 1 UUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAAGGCCACCGCCGACUAGAGAAUCAAUAAUUCUUUUGGGCGCUGGAAUAGCAUUGGCUAAGCU----CUA-ACGA .....-.(((.(.((((((((((((........))))))))))))..(((((..(((.(..(((((((.....)))))))).)))((((...))))..)))))..).)----)).-.... ( -31.10) >DroYak_CAF1 21043 107 - 1 UUAAUUUAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GCUGCCGCAGACUAGAGAAUCAAUAAUUCUAUUGGGUGCUA-----------GCUAAGCUGGCGAUG-UCGA ........(((..((((((((((((........))))))))))))..-..((((((((.((((....((((((....))))))...)))-----------)))..)).))))..)-)).. ( -29.40) >consensus UUAAU_CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA_GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUUUGGGUGCUGGAAUAGCAAUGCCUAAGCUAGCUCUG_UCGA ......((((((.((((((((((((........))))))))))))..............(.(((((((.....))))))).)))).)))............................... (-20.20 = -20.12 + -0.08)


| Location | 7,484,564 – 7,484,676 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7484564 112 - 22224390 GCCUAAUUAAAUAUU------AUGUGAAUCCUUUCUGGGCUUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGGGAAUCAAUAAUUCUUU ............(((------((.(((.((((((((((((((...-.......((((((((((((........)))))))))))).)-)))))...)))..))))).))))))))..... ( -26.40) >DroSec_CAF1 17633 112 - 1 ACCUAAUUAAAUAUU------AUGUGAAUCUUUUCUGGGCUUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUU ............(((------((.(((.((((((((((((((...-.......((((((((((((........)))))))))))).)-)))))...)))..))))).))))))))..... ( -24.40) >DroSim_CAF1 19318 112 - 1 ACCUAAUUAAAUAUU------AUGUGAAUCCUUUCUGGGCUUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUU ...............------.((((...(((((((((......)-))))...((((((((((((........))))))))))))))-))...))))....(((((((.....))))))) ( -24.30) >DroEre_CAF1 18637 113 - 1 GCCUAAUUAAAUACU------AUGUGAAUCCUUUUUGGCCUUAAU-CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAAGGCCACCGCCGACUAGAGAAUCAAUAAUUCUUU ...............------..............((((((((..-........(((((((((((........))))))))))))))))))).........(((((((.....))))))) ( -23.70) >DroYak_CAF1 21071 119 - 1 GCCUAAUUAAAUAGUAGUAGCAUGUGAAUCCUUUUUGGCCUUAAUUUAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA-GCUGCCGCAGACUAGAGAAUCAAUAAUUCUAU .............(..(((((..............(((.((......)).)))((((((((((((........))))))))))))..-)))))..).......(((((.....))))).. ( -26.50) >consensus GCCUAAUUAAAUAUU______AUGUGAAUCCUUUCUGGGCUUAAU_CAGACCAAAGCAUUAAUAUAAAUGAACGUAUUAAUGCUUAA_GGCCCCGCAGACUAGAGAAUCAAUAAUUCUUU ......................((((.........(((.((......)).)))((((((((((((........))))))))))))........))))....(((((((.....))))))) (-18.76 = -19.00 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:41 2006