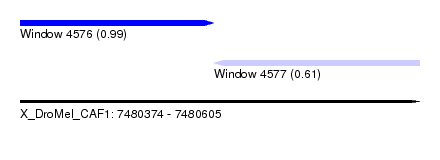
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,480,374 – 7,480,605 |
| Length | 231 |
| Max. P | 0.988156 |

| Location | 7,480,374 – 7,480,486 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7480374 112 + 22224390 -------CCCCACCCAAAUGUUGUGCGUAAAAACCGCAACAUUUGCAGACUCUGAA-ACCACCCACCUAUAUCCAUUUCCCCGAUUCUGCCUCUCACUCUCGUUUGGGCUGUAUUCUUUU -------.((((..(((((((((((.((....))))))))))))(((((.((.(((-(.................))))...)).)))))...........)..))))............ ( -23.43) >DroSec_CAF1 13515 111 + 1 -------CCCCACUCAAAUGUUGUGCGUAAACACCGCAACAUUUGCAGACUCUGA--ACCACCCACCUAUAUUCAUUUCCCCGAUUCUGCCUCUCACUCUCGUUUGGGCUGUAUUCUUUU -------.((((..(((((((((((.((....))))))))))))(((((.((...--.........................)).)))))...........)..))))............ ( -22.29) >DroSim_CAF1 13991 112 + 1 -------CCCCACCCAAAUGUUGUGCGUAAAAACCGCAACAUUUGCAGACUCUGAA-ACCACCCACCUAUAUCCAUUUCCCCGAUUCUGCCUCUCACUCUCGUUUGGGCUGUAUUCUUUU -------.((((..(((((((((((.((....))))))))))))(((((.((.(((-(.................))))...)).)))))...........)..))))............ ( -23.43) >DroEre_CAF1 14447 118 + 1 CCACCCGCCACGCCCCAAUGUUGUGCGUAAAAACCGCAACAUUUGCAGACUCUGAAAACCACCCACCUAUAUCCAUUUCCCCGAUUCUGCCUCUCACUCUCGACUG-GCCGUAUGCUU-U ......((.(((((..(((((((((.((....))))))))))).(((((.((.((((..................))))...)).)))))...............)-).)))..))..-. ( -21.47) >consensus _______CCCCACCCAAAUGUUGUGCGUAAAAACCGCAACAUUUGCAGACUCUGAA_ACCACCCACCUAUAUCCAUUUCCCCGAUUCUGCCUCUCACUCUCGUUUGGGCUGUAUUCUUUU ........((((..(((((((((((.((....))))))))))))(((((.((..............................)).)))))...........)..))))............ (-19.45 = -19.76 + 0.31)

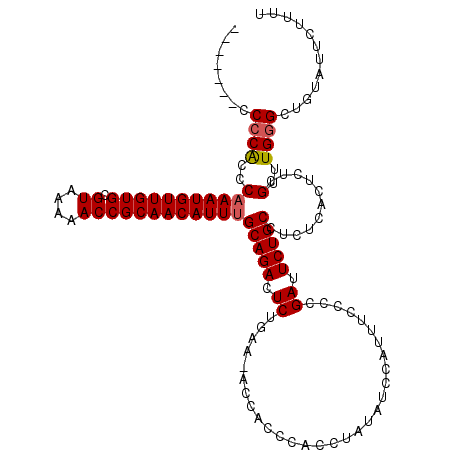
| Location | 7,480,486 – 7,480,605 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.605041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7480486 119 - 22224390 GCUCUCAAUCCUGGGCUUAGUUUUGACCUGAUUUCGAGACGGUUUAGCCGACUUAAAGCCACUUGGC-AAAAACAAAAAAAAACAAAACCCUCGAAAAAAUUCGCGGAUAAACAAAGGAA ........((((..(.(((((((((.......((..((.(((.....))).))..))(((....)))-...............))))))((.((((....)))).)).))).)..)))). ( -25.90) >DroSec_CAF1 13626 109 - 1 GCUCUCAAUCCUGGGCUUAGUUUUGACCUGAUUUCGAGACUGUUUAGCCGACUUAAAGCCACUUGGC-AAAAA----------AAAACCCCUCGAAAAAAUUCGCGGAUAAACAAAGGAA ........((((.(((((((((((((.......)))))))))...))))........(((....)))-.....----------......((.((((....)))).))........)))). ( -26.70) >DroSim_CAF1 14103 111 - 1 GCUCUCAAUCCUGGGCUUAGUUUUGACCUGAUUUCGAGACUGUUUAGCCGACUUAAAGCCACUUGGC-AAAAAA--------AAAAACCCCUCGAAAAAAUUCGCGGAUAAACAAAGGAA ........((((.(((((((((((((.......)))))))))...))))........(((....)))-......--------.......((.((((....)))).))........)))). ( -26.70) >DroEre_CAF1 14565 114 - 1 GCUCCCAAUCCUGCGCUUAGUUUUGACCUGAUUUCGAGGCUGUUUAGCCGACUUAAAGCCACUUGGGCAAAAA------AAAGAAAACCCCUCGAAAAAAUUCGCGGAUAAACAAAGGAA .......((((.(((.....((((((...(.((((..((((....))))........(((.....))).....------...)))).)...)))))).....)))))))........... ( -23.90) >consensus GCUCUCAAUCCUGGGCUUAGUUUUGACCUGAUUUCGAGACUGUUUAGCCGACUUAAAGCCACUUGGC_AAAAA_________AAAAACCCCUCGAAAAAAUUCGCGGAUAAACAAAGGAA ........((((.(((((((((((((.......)))))))))...))))........(((....)))......................((.((((....)))).))........)))). (-21.14 = -21.70 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:33 2006