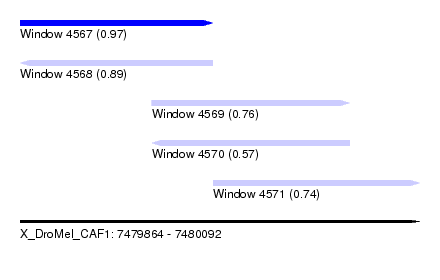
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,479,864 – 7,480,092 |
| Length | 228 |
| Max. P | 0.972742 |

| Location | 7,479,864 – 7,479,974 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 98.21 |
| Mean single sequence MFE | -44.00 |
| Consensus MFE | -40.70 |
| Energy contribution | -41.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479864 110 + 22224390 AAUUGCCCGCCCCCUUUUUGCGAAAAUACGCGCAACCUGCGCAUGUUGUUGACAUUGCGUUGAGUGUGGACAAUGUGUGUG--AGUGGGUGUUGGGUGUGUGAAAAGUGGGU ........((((.(((((..((.....((((.((.(..((((((((((((.(((((......))))).)))))))))))).--.))).))))......))..))))).)))) ( -43.20) >DroSec_CAF1 13011 112 + 1 AAUUGCCCGCCCCCUUUUUGCGAAAAUACGCGCAACCUGCGCAUGUUGUUGACAUUGCGUUGAGUGUGGACAAUGUGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGU ........((((.(((((..((.....((((.((..((((((((((((((.(((((......))))).)))))))))))).))..)).))))......))..))))).)))) ( -43.90) >DroSim_CAF1 13492 112 + 1 AAUUGCCCGCCCCCUUUUUGGGAAAAUACGCGCAACCUGCGCAUGUUGUUGACAUUGCGUUGAGUGUGGACAAUGUGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGU ....((((((.(((.....)))...((((((.((((((((((((((((((.(((((......))))).)))))))))))).......)).)))).)))))).....)))))) ( -44.91) >consensus AAUUGCCCGCCCCCUUUUUGCGAAAAUACGCGCAACCUGCGCAUGUUGUUGACAUUGCGUUGAGUGUGGACAAUGUGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGU ........((((.(((((((((..(((((.(((....(((((((((((((.(((((......))))).)))))))))))))...))).))))).....))))))))).)))) (-40.70 = -41.03 + 0.33)


| Location | 7,479,864 – 7,479,974 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 98.21 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479864 110 - 22224390 ACCCACUUUUCACACACCCAACACCCACU--CACACACAUUGUCCACACUCAACGCAAUGUCAACAACAUGCGCAGGUUGCGCGUAUUUUCGCAAAAAGGGGGCGGGCAAUU ................(((..(.(((...--.....(((((((...........))))))).......((((((.....)))))).............))))..)))..... ( -23.80) >DroSec_CAF1 13011 112 - 1 ACCCACUUUUCACACACCCAACACCCACUCUCACACACAUUGUCCACACUCAACGCAAUGUCAACAACAUGCGCAGGUUGCGCGUAUUUUCGCAAAAAGGGGGCGGGCAAUU .......................(((.(((((....(((((((...........))))))).......((((((.....)))))).............))))).)))..... ( -25.40) >DroSim_CAF1 13492 112 - 1 ACCCACUUUUCACACACCCAACACCCACUCUCACACACAUUGUCCACACUCAACGCAAUGUCAACAACAUGCGCAGGUUGCGCGUAUUUUCCCAAAAAGGGGGCGGGCAAUU .......................(((.(((((....(((((((...........))))))).......((((((.....)))))).............))))).)))..... ( -25.40) >consensus ACCCACUUUUCACACACCCAACACCCACUCUCACACACAUUGUCCACACUCAACGCAAUGUCAACAACAUGCGCAGGUUGCGCGUAUUUUCGCAAAAAGGGGGCGGGCAAUU ................(((..(.(((..........(((((((...........))))))).......((((((.....)))))).............))))..)))..... (-23.80 = -23.80 + 0.00)


| Location | 7,479,939 – 7,480,052 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -42.63 |
| Consensus MFE | -35.83 |
| Energy contribution | -39.50 |
| Covariance contribution | 3.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479939 113 + 22224390 UGUGUG--AGUGGGUGUUGGGUGUGUGAAAAGUGGGUGGCCUGUGGA-AUGGGUUGAUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGG ..((((--((...((((..(.(((.(....)...(((((((......-.((((..((((....)))).))))..)))))))))).)..))))...)))))).(....)........ ( -42.10) >DroSec_CAF1 13086 115 + 1 UGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGUGGCCUGUGGA-AUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGG ..(((((((....((((..(.(((.(....)...(((((((......-.(((((((.((....)))))))))..)))))))))).)..))))..))))))).(....)........ ( -43.20) >DroSim_CAF1 13567 116 + 1 UGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGUGGCCUGUGGAAAUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGG ..(((((((....((((..(.(((.(....)...(((((((........(((((((.((....)))))))))..)))))))))).)..))))..))))))).(....)........ ( -42.60) >consensus UGUGUGAGAGUGGGUGUUGGGUGUGUGAAAAGUGGGUGGCCUGUGGA_AUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGG ............(((.(((..(((((.((...(((((((((........(((((((.((....)))))))))..))))))).)).)).))))).((((.....))))))).))).. (-35.83 = -39.50 + 3.67)


| Location | 7,479,939 – 7,480,052 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.39 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479939 113 - 22224390 CCGGCCUUGUCUCCCUGCGAAAUUGUGUCAAUUGCAGUGGCCACUGGGAGAUGUCACCAUCAACCCAU-UCCACAGGCCACCCACUUUUCACACACCCAACACCCACU--CACACA ..(((((.((..((((((((...........)))))).))..))((((.((((....))))..)))).-.....))))).............................--...... ( -25.10) >DroSec_CAF1 13086 115 - 1 CCGGCCUUGUCUCCCUGCGAAAUUGUGUCAAUUGCAGUGGCCACUGGGAGAUGUCACCAGCCCCCCAU-UCCACAGGCCACCCACUUUUCACACACCCAACACCCACUCUCACACA ..(((((.(((((((((.(..((((((.....))))))..)))..)))))))((.....)).......-.....)))))..................................... ( -23.00) >DroSim_CAF1 13567 116 - 1 CCGGCCUUGUCUCCCUGCGAAAUUGUGUCAAUUGCAGUGGCCACUGGGAGAUGUCACCAGCCCCCCAUUUCCACAGGCCACCCACUUUUCACACACCCAACACCCACUCUCACACA ..((..((((......))))...(((((.((..(..((((((....(((((((............)))))))...))))))...)..)).))))).)).................. ( -25.00) >consensus CCGGCCUUGUCUCCCUGCGAAAUUGUGUCAAUUGCAGUGGCCACUGGGAGAUGUCACCAGCCCCCCAU_UCCACAGGCCACCCACUUUUCACACACCCAACACCCACUCUCACACA ..(((((((((((((((.(..((((((.....))))))..)))..)))))))).....................)))))..................................... (-22.39 = -22.39 + 0.00)

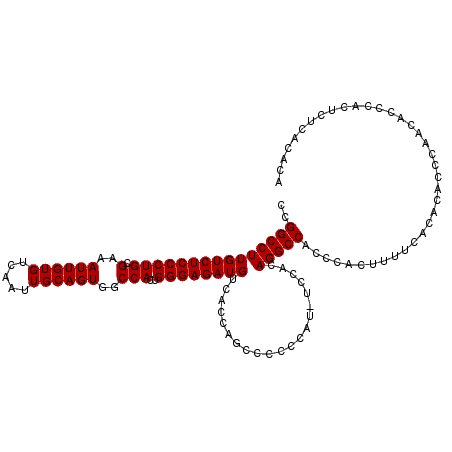
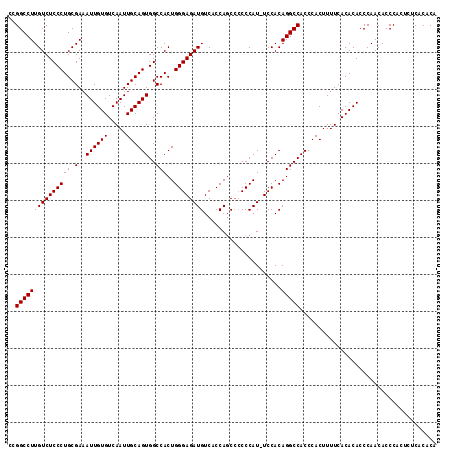
| Location | 7,479,974 – 7,480,092 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -46.33 |
| Consensus MFE | -36.46 |
| Energy contribution | -40.03 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479974 118 + 22224390 GGCCUGUGGA--AUGGGUUGAUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGGCAGUCAGUCGGCUGGCAUUGACCAAAAGGCACUCGCACUC .((((.(((.--.((((..((((....)))).))))..((((.(((((((((...)))))..))))(....)..))))(.(((((....))))).).....)))..)))).......... ( -46.90) >DroSec_CAF1 13123 114 + 1 GGCCUGUGGA--AUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGGCAGUCAGUCGGCUGGCAUUGACCAAAAGGCACUCGC---- .((((.(((.--.(((((((.((....)))))))))..((((.(((((((((...)))))..))))(....)..))))(.(((((....))))).).....)))..))))......---- ( -47.30) >DroSim_CAF1 13604 115 + 1 GGCCUGUGGAA-AUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGGCAGUCAGUCGGCUGGCAUUGACCAAAAGGCACUCGC---- .((((.(((..-.(((((((.((....)))))))))..((((.(((((((((...)))))..))))(....)..))))(.(((((....))))).).....)))..))))......---- ( -47.30) >DroEre_CAF1 14022 113 + 1 GA---GUGAAAAGUGGGGGUCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGGCAGUCAGUCGGCUGGCAUUGACCAAAAGGCACUCGC---- ((---(((...(.((((((..((....)).)))))).)((((.(((((((((...)))))..))))(....)..))))(((((((((....)))))..)).))......)))))..---- ( -43.80) >consensus GGCCUGUGGA__AUGGGGGGCUGGUGACAUCUCCCAGUGGCCACUGCAAUUGACACAAUUUCGCAGGGAGACAAGGCCGGCAGUCAGUCGGCUGGCAUUGACCAAAAGGCACUCGC____ .....((((....(((((((.((....)))))))))((((((.(((((((((...)))))..))))(....)..(((((((.....)))))))))).....((....))))))))).... (-36.46 = -40.03 + 3.56)

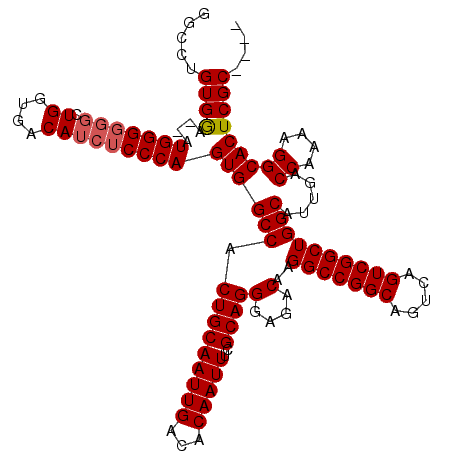
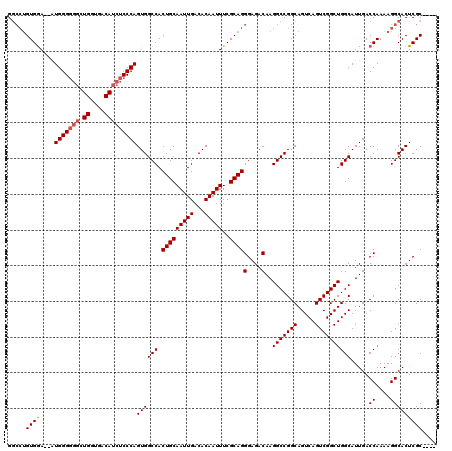
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:28 2006