
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,479,410 – 7,479,589 |
| Length | 179 |
| Max. P | 0.986614 |

| Location | 7,479,410 – 7,479,512 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -23.83 |
| Energy contribution | -23.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
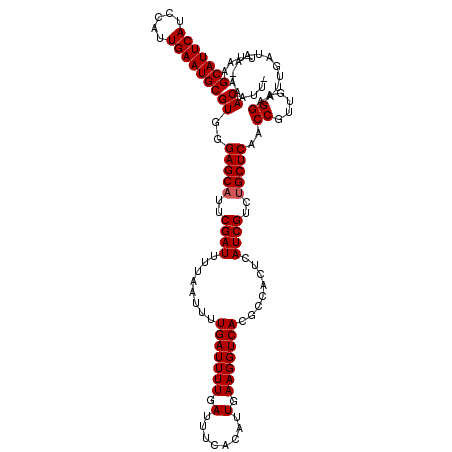
>X_DroMel_CAF1 7479410 102 + 22224390 UAAGCGUUCGCCUCGUUUCGUUUCGGUUUUGUCGUAUCUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUU .....((((.((.(...............((..(.((((((((......)))))))))..))....(((((((.....)))))))).))))))......... ( -25.20) >DroSec_CAF1 12573 102 + 1 UAAGCGUUCGCCUCGCUUCGUUUCGGUUUUGUCGUAUUUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUU ....((...(((((((.............((..(.((((((((......)))))))))..))....(((((((.....)))))))))))).))...)).... ( -24.30) >DroSim_CAF1 12600 102 + 1 UAAGCGUUCGCCUCGCUUCGUUUUGGUUUUGUCGUAUUUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUU ....((...(((((((.............((..(.((((((((......)))))))))..))....(((((((.....)))))))))))).))...)).... ( -24.30) >consensus UAAGCGUUCGCCUCGCUUCGUUUCGGUUUUGUCGUAUUUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUU .(((((.......))))).(((((.....((..(.((((((((......)))))))))..))....(((((((.....)))))))...)))))......... (-23.83 = -23.17 + -0.66)

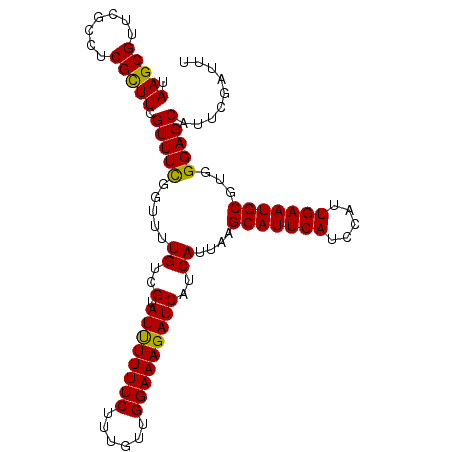
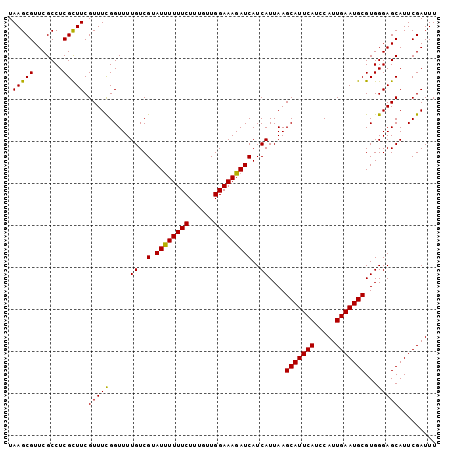
| Location | 7,479,447 – 7,479,552 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 98.73 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -27.20 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479447 105 + 22224390 CUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCAUGCCACUC ...........(((...((((.(((....(((((((.....)))))))(((..(.((.((((........))))...)).)..)))..))).))))...)))... ( -27.20) >DroSec_CAF1 12610 105 + 1 UUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUC ...........(((...((((.(((....(((((((.....)))))))(((..(.((.((((........))))...)).)..)))..))).))))...)))... ( -27.20) >DroSim_CAF1 12637 105 + 1 UUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUC ...........(((...((((.(((....(((((((.....)))))))(((..(.((.((((........))))...)).)..)))..))).))))...)))... ( -27.20) >consensus UUUUUCUUUGUUGGAAAGAUCAUCAUUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUC ...........(((...((((.(((....(((((((.....)))))))(((..(.((.((((........))))...)).)..)))..))).))))...)))... (-27.20 = -27.20 + -0.00)

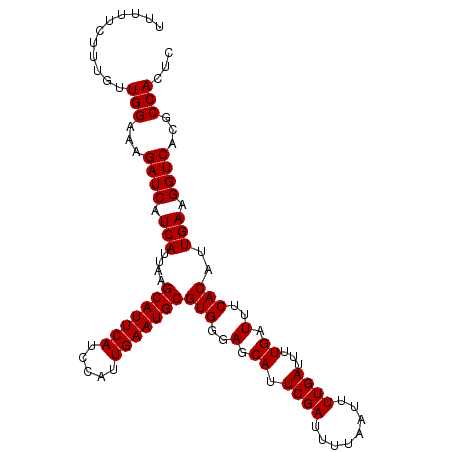
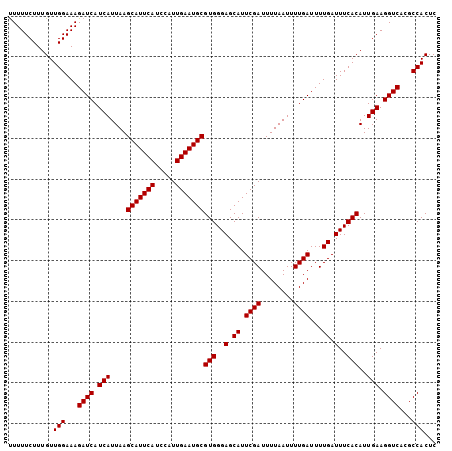
| Location | 7,479,472 – 7,479,589 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -28.29 |
| Consensus MFE | -25.86 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7479472 117 + 22224390 UUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCAUGCCACUCAUCGUCUGCUCAACCGUUGAGGAAUUGAUAA-AUACAU-- ....(((((((.....)))))))(((.(((((..((((........(((((((.(........).)))))))........))))..)))))..((.....)).........-.)))..-- ( -28.99) >DroSec_CAF1 12635 118 + 1 UUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUCAUCGUCUGCUCAACCGUUGAGGAAUUGAUAA-AAACAUU- ....(((((((.....)))))))(((.(((((..((((........(((((((.(........).)))))))........))))..)))))..((.....)).........-...))).- ( -27.69) >DroSim_CAF1 12662 119 + 1 UUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUCAUCGUCUGCUCAACCGUUGAGGAAUUGAUAA-AAACAUUU ....(((((((.....)))))))(((.(((((..((((........(((((((.(........).)))))))........))))..)))))..((.....)).........-...))).. ( -27.69) >DroEre_CAF1 13519 120 + 1 UUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCGCAUUGAAGGUCACGCCACUCAUCGACCGCUCGCCCGCCGAGGAAUUGAUAAGAAACAAAC ....(((((((.....)))))))((((((((..(((((........(((((((.(........).)))))))........)))))..)))..)))))....................... ( -28.79) >consensus UUAAGCAUUCAUCCAUUGAAUGCGUGGGAGCAUUCGAUUUUAAUUUUGAUUUUGAUUUCACAUUGAAGGUCACGCCACUCAUCGUCUGCUCAACCGUUGAGGAAUUGAUAA_AAACAUU_ ....(((((((.....)))))))((..(((((..((((........(((((((.(........).)))))))........))))..)))))..((.....))............)).... (-25.86 = -26.12 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:24 2006