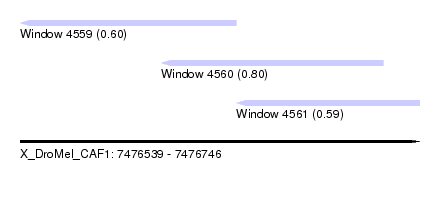
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,476,539 – 7,476,746 |
| Length | 207 |
| Max. P | 0.795309 |

| Location | 7,476,539 – 7,476,651 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -26.84 |
| Energy contribution | -26.52 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7476539 112 - 22224390 AGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCAU-----AUCUGCC--UGACGAGCUCGUAAUAUAGCCAUAGAUAUAGCCGAGAUAUCAGUUAAUCGGUGGGCCAUAUAGUC ..(((.(((.((-(........((((......(((((....-----..)))))--))))((.((((...((((........))))..))))...))......))).))))))........ ( -30.50) >DroSec_CAF1 9751 112 - 1 AGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCAU-----AUCUGCC--UGACGAGCUCGAAAUAUAGCCAUAGAUAUAGCCGAGAUAUCAGUUAAUCGGUGGGCCAUAUAGUC ..(((.(((.((-(........((((......(((((....-----..)))))--))))((.((((...((((........))))..))))...))......))).))))))........ ( -30.50) >DroSim_CAF1 9301 112 - 1 AGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCAU-----AUCUGCC--UGACGAGCUCGAAAUAUAGCCAUAGAUAUAGCCGAGAUAUCAGUUAAUCGGUGGGCCAUAUAGUC ..(((.(((.((-(........((((......(((((....-----..)))))--))))((.((((...((((........))))..))))...))......))).))))))........ ( -30.50) >DroEre_CAF1 10486 114 - 1 AGGGUGCCAGCA-AGAUAAGAAGUCAAAAAGUGGCAGUCAU-----AUCUGCCUCUGACGAGCUCGAAAUAUAGCCAUAGAUAUAGUCGAGAUAUCAGUUAAUCGGUGGGCCAUAUAGUC ..(((.(((.(.-.........((((....(.(((((....-----..))))).)))))((.(((((..((((........)))).)))))...))........).))))))........ ( -29.30) >DroYak_CAF1 13802 118 - 1 AGGGUGCCAGCAAAGAUAAGAAGUCAAGAAGUGGCAGUCAUACAGUAUCUGCC--UGACGAGCUCGAAAUAUGGCCAUAGAUAUAGCCGAGAUAUCAGUUAAUCGGUGGACCAUAUACAC .((((((((.....(((.....)))......)))).((((..(((...)))..--))))..))))...((((((((((.(((.((((.((....)).))))))).)))).)))))).... ( -32.00) >consensus AGGGUGCCAGCG_AGAUAAGAAGUCAAGAAGUGGCAGUCAU_____AUCUGCC__UGACGAGCUCGAAAUAUAGCCAUAGAUAUAGCCGAGAUAUCAGUUAAUCGGUGGGCCAUAUAGUC ..(((.(((.(...........((((......(((((...........)))))..))))((.((((...((((........))))..))))...))........).))))))........ (-26.84 = -26.52 + -0.32)



| Location | 7,476,612 – 7,476,727 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -19.06 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7476612 115 - 22224390 AGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAAGAAAUUACAACAA---AAAGCGAGACAAGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCA ..........(((((..(((...((......))...)-))(((...............))).....---...)))))....((.(((((.(.-.(((.....)))..)...))))).)). ( -23.26) >DroSec_CAF1 9824 115 - 1 AGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAACAAAUUACAACAA---AAAGCGAGACAAGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCA ..........(((((..(((...((......))...)-))(((...............))).....---...)))))....((.(((((.(.-.(((.....)))..)...))))).)). ( -23.26) >DroSim_CAF1 9374 115 - 1 AGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAACAAAUUACAACAA---AAAGCGAGACAAGGGUGCCAGCG-AGAUAAGAAGUCAAGAAGUGGCAGUCA ..........(((((..(((...((......))...)-))(((...............))).....---...)))))....((.(((((.(.-.(((.....)))..)...))))).)). ( -23.26) >DroEre_CAF1 10561 119 - 1 GGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUACGCGUAUAAAUAAAAACAAAUUACAACAAAAAAAAGCGAGACGAGGGUGCCAGCA-AGAUAAGAAGUCAAAAAGUGGCAGUCA (((((....(((((.((((((.(((............)))(((...............))).........)))).)).))))))))))....-.........((((.....))))..... ( -22.16) >DroYak_CAF1 13880 117 - 1 AGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUACGCGUAUAAAUAAAAGCAAAUUACAACAA---AAAGCGAGACGAGGGUGCCAGCAAAGAUAAGAAGUCAAGAAGUGGCAGUCA .......(((((((.((((((.(((............)))(((...............))).....---.)))).)).)))))))((((.....(((.....)))......))))..... ( -22.36) >consensus AGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA_GCGUAUAAAUAAAAACAAAUUACAACAA___AAAGCGAGACAAGGGUGCCAGCG_AGAUAAGAAGUCAAGAAGUGGCAGUCA ..........(((((..((....((......)).....))(((...............)))...........)))))....((.(((((.....(((.....)))......))))).)). (-19.06 = -19.06 + 0.00)

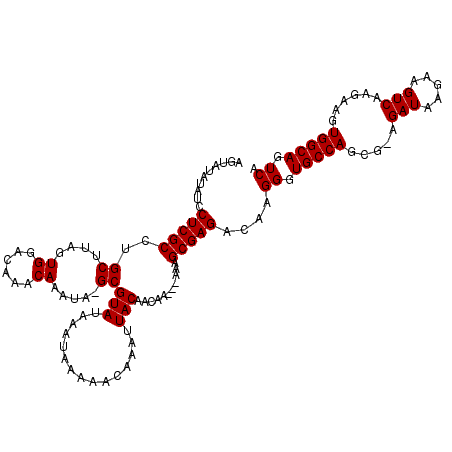
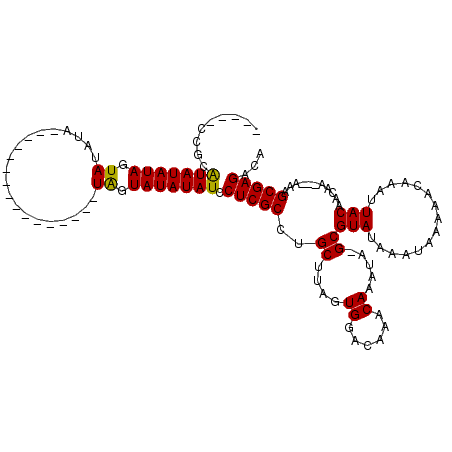
| Location | 7,476,651 – 7,476,746 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.42 |
| Mean single sequence MFE | -15.84 |
| Consensus MFE | -11.53 |
| Energy contribution | -11.13 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7476651 95 - 22224390 -----CCGCAUAUAUAGUAUAUA----------------UAGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAAGAAAUUACAACAA---AAAGCGAGACA -----....(((((((.((....----------------)).))))))).(((((..(((...((......))...)-))(((...............))).....---...)))))... ( -15.96) >DroSec_CAF1 9863 95 - 1 -----CCGCGUAUAUAGUAUAUA----------------UAGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAACAAAUUACAACAA---AAAGCGAGACA -----....(((((((.((....----------------)).))))))).(((((..(((...((......))...)-))(((...............))).....---...)))))... ( -16.26) >DroSim_CAF1 9413 95 - 1 -----CCGCGUAUAUAGUAUAUA----------------UAGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA-GCGUAUAAAUAAAAACAAAUUACAACAA---AAAGCGAGACA -----....(((((((.((....----------------)).))))))).(((((..(((...((......))...)-))(((...............))).....---...)))))... ( -16.26) >DroEre_CAF1 10600 99 - 1 -----GCACAUAUAUAGUAUAAA----------------UGGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUACGCGUAUAAAUAAAAACAAAUUACAACAAAAAAAAGCGAGACG -----....(((((((.((....----------------)).))))))).(((((..((....((......)).....))(((...............)))...........)))))... ( -12.66) >DroYak_CAF1 13920 117 - 1 AAAACACACAUAUAUAGUAUAUAUAUAGUAGAUAUAGUAUAGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUACGCGUAUAAAUAAAAGCAAAUUACAACAA---AAAGCGAGACG .........(((((((.(((((.((((.....))))))))).))))))).(((((..((....((......)).....))(((...............))).....---...)))))... ( -18.06) >consensus _____CCGCAUAUAUAGUAUAUA________________UAGUAUAUAUCCUCGCCUGCUUAGUGGACAAACAAAUA_GCGUAUAAAUAAAAACAAAUUACAACAA___AAAGCGAGACA .........(((((((.((....................)).))))))).(((((..((....((......)).....))(((...............)))...........)))))... (-11.53 = -11.13 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:20 2006