
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,469,381 – 7,469,652 |
| Length | 271 |
| Max. P | 0.994617 |

| Location | 7,469,381 – 7,469,482 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.35 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -21.74 |
| Energy contribution | -21.58 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469381 101 + 22224390 AGUCG--AGUCCCAGCUUCAU--AGCCACUCAAGUAACCGAGUUACGUGAGGUUAC-UCCAUACGGGGCAG---CAA-----------AAAUAGGGGGCCAGGAUGUGGGGAAGUGAAGA ..(((--..((((.((.((..--.(((.((((.(((((...))))).))))(((.(-(((....)))).))---)..-----------........)))...)).)).))))..)))... ( -33.30) >DroSec_CAF1 2641 101 + 1 AGUCA--AGUCCCAGCUUCAU--AGCCACUCAAGUAACCGAGUUACGUGAGGUUAC-UCCAUACGGGGCAG---CCA-----------AAAGAGGAGGCCAGGAUGUGGGGAAGUGAAGA ..(((--..((((.((.((..--.(((.(((..(((((...)))))....((((.(-(((....)))).))---)).-----------...)))..)))...)).)).))))..)))... ( -36.00) >DroSim_CAF1 4348 101 + 1 AGUCG--AGUCCCAGCUUCAU--AGCCACUCAAGUAACCGAGUUACGUGAGGUUAC-UCCAUACGGGGCAG---CCA-----------AAAGAGGAGGCCAGGAUGUGGGGAAGUGAAGA ..(((--..((((.((.((..--.(((.(((..(((((...)))))....((((.(-(((....)))).))---)).-----------...)))..)))...)).)).))))..)))... ( -35.30) >DroEre_CAF1 2950 103 + 1 UAGUA--AGUCCCAGCUUCAUACAGCCACUCAAGUAAGCGAGUUACGUGAGGUUAC-UUCAUAGGGGGAGG---CCA-----------GGAGGGGGGGCCAGGAUGUGGGGAAGUGAAGA .....--.......(((((.((((.((.((((.((((.....)))).))))....(-(((....)))).((---((.-----------........)))).)).))))..)))))..... ( -26.30) >DroYak_CAF1 4884 118 + 1 AGUCAAGAGUCCCAGCUUCAU--AGCCACUCAAGUAAGCGAGUUACGUGAGGUUGCCUCCAUAAGGGGGGGUUGCCAGCAAAAAAUAAGAAAUGGGGGCCAGGAUAUGGGGAAGUGAAGA ..(((....(((((...((..--.(((.((((.((((.....))))((..(((.((((((......)))))).))).)).............)))))))...))..)))))...)))... ( -37.20) >consensus AGUCA__AGUCCCAGCUUCAU__AGCCACUCAAGUAACCGAGUUACGUGAGGUUAC_UCCAUACGGGGCAG___CCA___________AAAGAGGGGGCCAGGAUGUGGGGAAGUGAAGA ..(((....((((.((.((.....(((.((((.((((.....)))).))))......(((....))).............................)))...)).)).))))..)))... (-21.74 = -21.58 + -0.16)

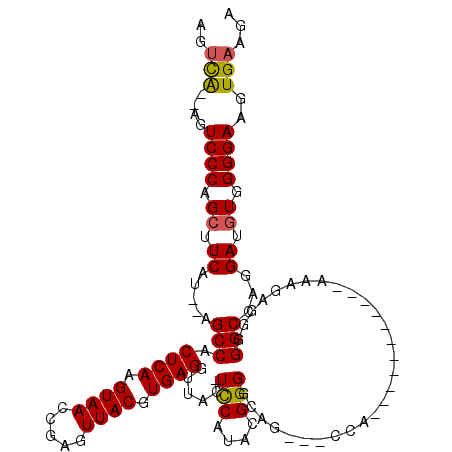
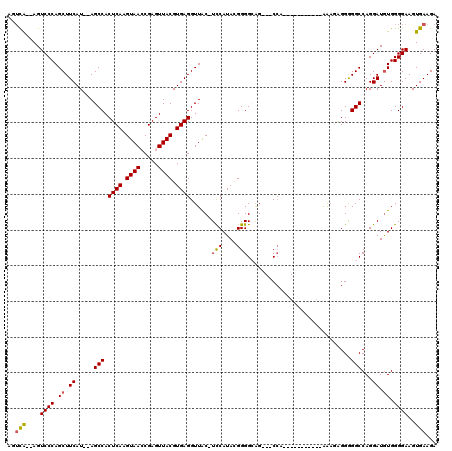
| Location | 7,469,450 – 7,469,562 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469450 112 - 22224390 CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCGUAUCUUCACUUCCCCACAUCCUGGCCCCCUAUUU-------- .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....)))..........-------- ( -20.32) >DroSec_CAF1 2710 112 - 1 CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUAUCUUCACUUCCCCACAUCCUGGCCUCCUCUUU-------- .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....)))..........-------- ( -20.32) >DroSim_CAF1 4417 112 - 1 CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUAUCUUCACUUCCCCACAUCCUGGCCUCCUCUUU-------- .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....)))..........-------- ( -20.32) >DroEre_CAF1 3021 112 - 1 CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUAUCUUCACUUCCCCACAUCCUGGCCCCCCCUCC-------- .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....)))..........-------- ( -20.32) >DroYak_CAF1 4962 120 - 1 CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUAUCUUCACUUCCCCAUAUCCUGGCCCCCAUUUCUUAUUUUU .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....))).................. ( -20.42) >consensus CUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUAUCUUCACUUCCCCACAUCCUGGCCCCCUCUUU________ .......(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))((.(((.............))).))......(((.....))).................. (-20.34 = -20.34 + -0.00)

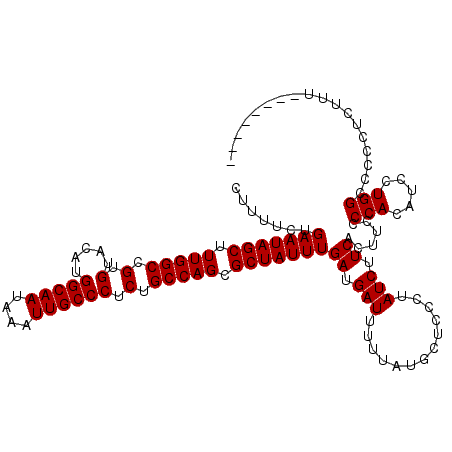

| Location | 7,469,482 – 7,469,602 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -30.36 |
| Energy contribution | -30.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469482 120 + 22224390 UACGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... ( -30.60) >DroSec_CAF1 2742 120 + 1 UAGGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... ( -30.30) >DroSim_CAF1 4449 120 + 1 UAGGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... ( -30.30) >DroEre_CAF1 3053 120 + 1 UAGGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... ( -30.30) >DroYak_CAF1 5002 120 + 1 UAGGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... ( -30.30) >consensus UAGGGAGCAUAAAAAUCAUCAAAUAGCGCUGGCAGAGGGCAAUUUAUUGCCAUGUACACGGCCAAAGCUAUUCAGAAAAGUGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAA ...(((((((.....((....((((((..((((....(((((....))))).((....))))))..))))))..))...))))))).((.((.(((.........))).))))....... (-30.36 = -30.36 + -0.00)


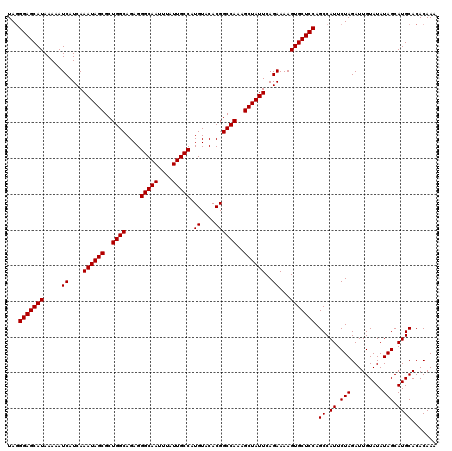
| Location | 7,469,482 – 7,469,602 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469482 120 - 22224390 UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCGUA ......(((..(((((............))))).((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))))) ( -32.40) >DroSec_CAF1 2742 120 - 1 UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUA .(((((((.......)))))))............((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))... ( -31.40) >DroSim_CAF1 4449 120 - 1 UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUA .(((((((.......)))))))............((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))... ( -31.40) >DroEre_CAF1 3053 120 - 1 UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUA .(((((((.......)))))))............((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))... ( -31.40) >DroYak_CAF1 5002 120 - 1 UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUA .(((((((.......)))))))............((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))... ( -31.40) >consensus UUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUUGCCCUCUGCCAGCGCUAUUUGAUGAUUUUUAUGCUCCCUA .(((((((.......)))))))............((((((....((.(((((((.(((((.(.(.....(((((....)))))).).))))).)))))))...))......))))))... (-31.58 = -31.58 + 0.00)



| Location | 7,469,522 – 7,469,632 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -32.09 |
| Consensus MFE | -27.74 |
| Energy contribution | -27.55 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469522 110 - 22224390 CAUAGCAAUG--CAUAUAUAUGGCCAUAGAUAUUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUU .((((((.((--((((((.(((........))).)))))))))))))).......(((..(((((..((((.............))))..))))).....)))......... ( -31.32) >DroSec_CAF1 2782 110 - 1 CAUAGCAAUG--CAUAUAUAUGGCCAUAGAUAUUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUU .((((((.((--((((((.(((........))).)))))))))))))).......(((..(((((..((((.............))))..))))).....)))......... ( -31.32) >DroSim_CAF1 4489 110 - 1 CAUAGCAAUG--CAUAUAUAUGGCCAUAGAUAUUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUU .((((((.((--((((((.(((........))).)))))))))))))).......(((..(((((..((((.............))))..))))).....)))......... ( -31.32) >DroEre_CAF1 3093 112 - 1 CAUAGCAAUGCACAUAUAUAUGGCUAUAGAUAUUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUU .((((((.((((((((.((((........)))).)))))))))))))).......(((..(((((..((((.............))))..))))).....)))......... ( -34.42) >consensus CAUAGCAAUG__CAUAUAUAUGGCCAUAGAUAUUUGUGUGCAUGCUAUAUACAAUCUAGAAUGGCUGGAGCACUUUUCUGAAUAGCUUUGGCCGUGUACAUGGCAAUAAAUU ............(((.((((((((((.((.((((((.((((..(((((............)))))....)))).....)))))).)).)))))))))).))).......... (-27.74 = -27.55 + -0.19)


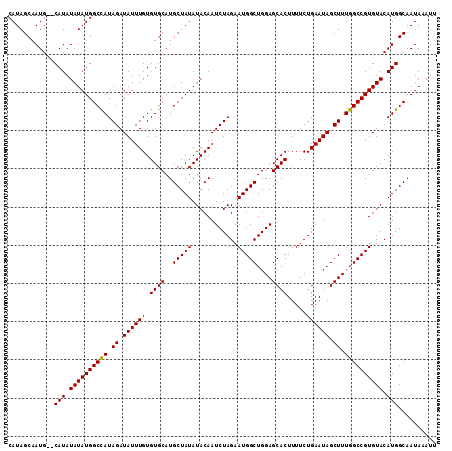
| Location | 7,469,562 – 7,469,652 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -19.99 |
| Energy contribution | -19.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7469562 90 + 22224390 UGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAAUAUCUAUGGCCAUAUAUAUG--CAUUGCUAUGUAUGGAUUGCUGGCCAUGUA---- ....(((((.((((......((((((((((((((.....((((........))))...))--)).)))))))))))))).))))).......---- ( -22.60) >DroSec_CAF1 2822 90 + 1 UGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAAUAUCUAUGGCCAUAUAUAUG--CAUUGCUAUGUAUGGAUUGCUGGCCAUGUA---- ....(((((.((((......((((((((((((((.....((((........))))...))--)).)))))))))))))).))))).......---- ( -22.60) >DroSim_CAF1 4529 90 + 1 UGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAAUAUCUAUGGCCAUAUAUAUG--CAUUGCUAUGUAUGGAUUGCUGGCCAUGUA---- ....(((((.((((......((((((((((((((.....((((........))))...))--)).)))))))))))))).))))).......---- ( -22.60) >DroEre_CAF1 3133 96 + 1 UGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAAUAUCUAUAGCCAUAUAUAUGUGCAUUGCUAUGUAUGGAUUGCUGGCCAUGUAUAUA (((.(((((.((((......((((((((((((((((.(.((((........)))).).)))))).)))))))))))))).)))))....))).... ( -28.70) >consensus UGCUCCAGCCAUUCUAGAUUGUAUAUAGCAUGCACACAAAUAUCUAUGGCCAUAUAUAUG__CAUUGCUAUGUAUGGAUUGCUGGCCAUGUA____ (((.(((((.((((......((((((((((.........((((.(((....))).))))......)))))))))))))).)))))....))).... (-19.99 = -19.98 + -0.00)

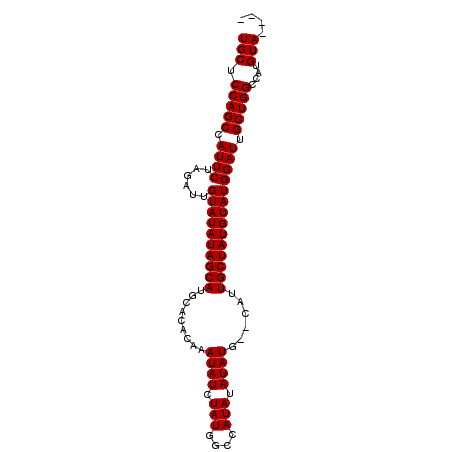

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:05 2006