
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,467,916 – 7,468,011 |
| Length | 95 |
| Max. P | 0.849293 |

| Location | 7,467,916 – 7,468,011 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7467916 95 + 22224390 -----------------------UGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGAUGCACUCAC--GCUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG -----------------------.(((((..((((((((((((.(((((....(((((.......))))).....)--)))))))).((.....))....)))))))).)))))...... ( -26.60) >DroSec_CAF1 1227 95 + 1 -----------------------UGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGAUGCACUCAC--GCUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG -----------------------.(((((..((((((((((((.(((((....(((((.......))))).....)--)))))))).((.....))....)))))))).)))))...... ( -26.60) >DroSim_CAF1 2928 95 + 1 -----------------------UGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGAUGCACUCAC--GCUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG -----------------------.(((((..((((((((((((.(((((....(((((.......))))).....)--)))))))).((.....))....)))))))).)))))...... ( -26.60) >DroEre_CAF1 1422 95 + 1 -----------------------UGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGUUGCACUCAC--GCUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG -----------------------.(((((..((((((((((((((.((((...(((....)))...)))).....)--))).((....))........)))))))))).)))))...... ( -23.00) >DroYak_CAF1 3369 120 + 1 GGUACUGGUACUGGUACUGAAACUGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGAUGCACUCACUCACUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG ((((((......))))))..(((.(((((..(((((((((((..(((.((...(((((.......)))))..)).)))....((....)).......))))))))))).))))))))... ( -29.70) >consensus _______________________UGGAGAAAUCAAAUUAAAGGCGGGCGAAACGCAUCAUUGUCAGAUGCACUCAC__GCUCCCUUAUGGCAUUCAAUUUUAAUUUGAAUUUCCGUUGAG ........................(((((..((((((((((((.((((((...(((((.......)))))..))....)))))))).((.....))....)))))))).)))))...... (-21.02 = -21.42 + 0.40)


| Location | 7,467,916 – 7,468,011 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -24.68 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
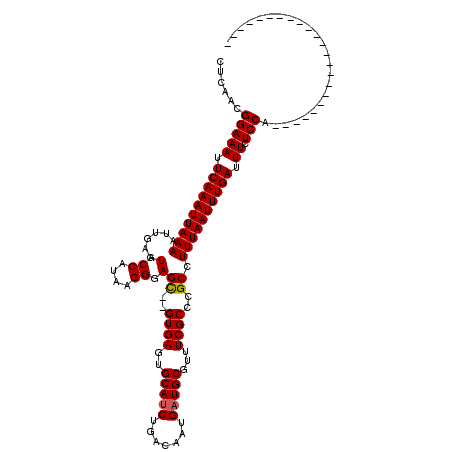
>X_DroMel_CAF1 7467916 95 - 22224390 CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGC--GUGAGUGCAUCUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCA----------------------- ......(((((.((((((((((..((.....))....((.((--(.(((((((((.......))))))))))))))...)))))))))).)).))).----------------------- ( -25.10) >DroSec_CAF1 1227 95 - 1 CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGC--GUGAGUGCAUCUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCA----------------------- ......(((((.((((((((((..((.....))....((.((--(.(((((((((.......))))))))))))))...)))))))))).)).))).----------------------- ( -25.10) >DroSim_CAF1 2928 95 - 1 CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGC--GUGAGUGCAUCUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCA----------------------- ......(((((.((((((((((..((.....))....((.((--(.(((((((((.......))))))))))))))...)))))))))).)).))).----------------------- ( -25.10) >DroEre_CAF1 1422 95 - 1 CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGC--GUGAGUGCAACUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCA----------------------- ......(((((.((((((((((..((.....))....((.((--(.(((((((.(.......).))))))))))))...)))))))))).)).))).----------------------- ( -20.50) >DroYak_CAF1 3369 120 - 1 CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGUGAGUGAGUGCAUCUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCAGUUUCAGUACCAGUACCAGUACC ......(((((.((((((((((......(.((....)).)(((.((((..(((((.......)))))...)))).))).)))))))))).)).))).......((((........)))). ( -27.60) >consensus CUCAACGGAAAUUCAAAUUAAAAUUGAAUGCCAUAAGGGAGC__GUGAGUGCAUCUGACAAUGAUGCGUUUCGCCCGCCUUUAAUUUGAUUUCUCCA_______________________ ......(((((.((((((((((......(.((....)).)((..((((..(((((.......)))))...))))..)).)))))))))).)).)))........................ (-21.26 = -21.30 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:54 2006