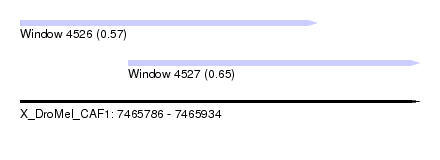
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,465,786 – 7,465,934 |
| Length | 148 |
| Max. P | 0.652799 |

| Location | 7,465,786 – 7,465,896 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -26.52 |
| Energy contribution | -27.52 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7465786 110 + 22224390 UGUCUGUCCGUCCCUCUCUGUCGCCACCGCAAAACGUGCGUAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGUUUAUACUGCAGCAUUCACAAAAGUCUUUAAAG---------- .....(..((((((((.((........((((.....))))........)).)))))).))..)(((((((((..((((....)))).)))))))))..............---------- ( -32.69) >DroSim_CAF1 809 109 + 1 UGUCUGUCCGUCCCUCUCUGUCGCCACCGCAAAACGUGCGUAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGU-UAUACUGUAGCAUUCACAAAAGUCUUUAAAG---------- .....(..((((((((.((........((((.....))))........)).)))))).))..)(((((((((.((((.-....)))))))))))))..............---------- ( -34.59) >DroYak_CAF1 940 117 + 1 UGUCUGUCUCUCUCUCGCUGUCGCCACCGCAAAACGUGCGUAAUUAAAAGUGAGGCAGUGUGCGUGAGUGUUUACACU-UAAAACGUGGCAUUCACAGAAAG--UUAAAGUUAAAUUUAA ..(((((......((((((........((((.....))))........))))))((..(((...((((((....))))-))..)))..))....)))))...--................ ( -28.29) >consensus UGUCUGUCCGUCCCUCUCUGUCGCCACCGCAAAACGUGCGUAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGU_UAUACUGUAGCAUUCACAAAAGUCUUUAAAG__________ .....(..((((((((.((........((((.....))))........)).)))))).))..)(((((((((.(((((....))))))))))))))........................ (-26.52 = -27.52 + 1.01)


| Location | 7,465,826 – 7,465,934 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7465826 108 + 22224390 UAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGUUUAUACUGCAGCAUUCACAAAAGUCUUUAAAG------------AAUUAAUCAUAAUUAUUUAUGUCGAAUUUUUGGACAAA .......................(((((((((..((((....)))).)))))))))....((((...(((------------((((...(((((....)))))...)))))))))))... ( -24.30) >DroSim_CAF1 849 107 + 1 UAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGU-UAUACUGUAGCAUUCACAAAAGUCUUUAAAG------------AAAUAAUCCUAAUUAUUUAAGACGAAUUUUUGGACAAA .........((..((((((...((((((((((.((((.-....)))))))))))))....(((((.....------------(((((((....))))))))))))).))))))..))... ( -24.90) >DroYak_CAF1 980 114 + 1 UAAUUAAAAGUGAGGCAGUGUGCGUGAGUGUUUACACU-UAAAACGUGGCAUUCACAGAAAG--UUAAAGUUAAAUUUAAAGUCCAAUUUUGAACUCAUUCUUUUGAGUUUU-G--CAAA .........(((((((..(((...((((((....))))-))..)))..)).))))).((...--(((((......)))))..)).......(((((((......))))))).-.--.... ( -25.70) >consensus UAAUUAAAAGUGAGGGAGUGUGCGUGAGUGUUUACAGU_UAUACUGUAGCAUUCACAAAAGUCUUUAAAG____________AAAUAAUCAUAAUUAUUUAUGUCGAAUUUUUGGACAAA ..........((((((.(....)(((((((((.(((((....)))))))))))))).....))))))..................................................... (-12.70 = -13.70 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:50 2006