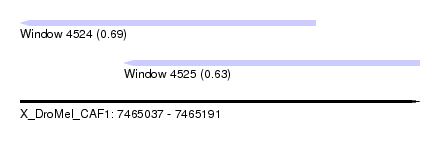
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,465,037 – 7,465,191 |
| Length | 154 |
| Max. P | 0.685557 |

| Location | 7,465,037 – 7,465,151 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7465037 114 - 22224390 AAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCCAAC--A----UGUAUGUGUGUGCUUAUCAGUGCUUGCCAAUUAUGAGGGA ((((((((((.......(((((......))))).))))))))))...((((((((...(((......))).((--(----(....))))(..((....))..).....)))))))).... ( -29.70) >DroSim_CAF1 73 112 - 1 AAUGUUUGGCAAUUAUGCGGAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCAAAC--A----UGUAUGU--GUGCUUAUCAGUGCUUGCCAAUUAUGAGGGA ((((((((((.......(((((......))))).))))))))))...((((((((...((.(((((((.....--.----.))))))--(..((....))..).).)))))))))).... ( -27.20) >DroYak_CAF1 81 118 - 1 AAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGCCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACUACAUAUAUGUAUGUAUGU--GUGCUUAUCAGUGCUUGCCAAUUAUGAGGGA (((((((((.......((.((((.....)))).)))))))))))...((((((((...((.(((((((((((....)))).))))))--(..((....))..).).)))))))))).... ( -29.61) >consensus AAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCAAAC__A____UGUAUGU__GUGCUUAUCAGUGCUUGCCAAUUAUGAGGGA ((((((((((.......(((((......))))).))))))))))...((((((((...((.(((((((.............))))))..(..((....))..).).)))))))))).... (-24.61 = -24.72 + 0.11)


| Location | 7,465,077 – 7,465,191 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7465077 114 - 22224390 UUUACCAGAAAACCCUCGAAACCCCCCGAUGGAUGCCAAAAAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCCAAC--A---- .......((...((.(((........))).))........((((((((((.......(((((......))))).))))))))))...)).........(((......)))...--.---- ( -24.90) >DroSim_CAF1 111 114 - 1 UUUACCAGAAAACCCUCGGAACCCCCCGACGGAUGCCAAAAAUGUUUGGCAAUUAUGCGGAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCAAAC--A---- ....((......((.((((......)))).))........((((((((((.......(((((......))))).))))))))))..............)).............--.---- ( -25.90) >DroYak_CAF1 119 108 - 1 UUUAGCAGAAAACCCUCGAA------------AUGCCAAAAAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGCCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACUACAUAUAUGUA ............((((..((------------.(((((((....))))))).....((.((((.....)))).))..................))..))))................... ( -19.50) >consensus UUUACCAGAAAACCCUCGAAACCCCCCGA_GGAUGCCAAAAAUGUUUGGCAAUUAUGCGAAGGGAAAACUUUGGGCCAAACAUUAUAUCAUAAUUAAAGGGAAUAUACCAAAC__A____ ............((((..................((((((....))))))((((((((((((......)))))(......).......)))))))..))))................... (-18.95 = -18.73 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:48 2006