
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,462,542 – 7,462,632 |
| Length | 90 |
| Max. P | 0.999822 |

| Location | 7,462,542 – 7,462,632 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -15.96 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.61 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
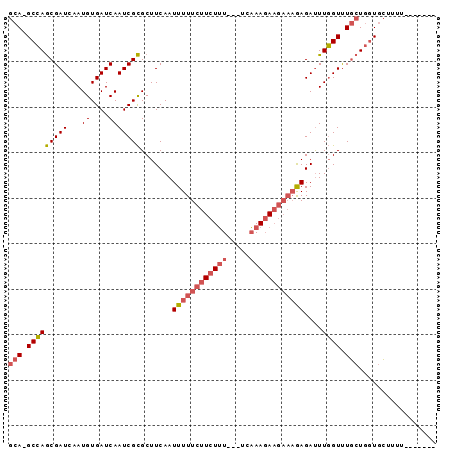
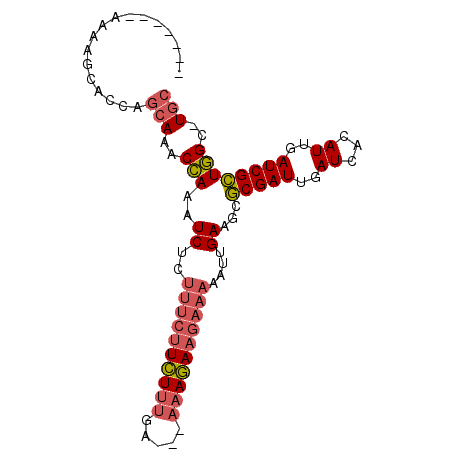
>X_DroMel_CAF1 7462542 90 + 22224390 GCA-GCCAGCGAUCAAUGUGAUCAAUCGCGCUUCAAUUUUUCUUCUUU---UCAAAGAAGAAAGAGAUUUGGUUUGCUGGUGCUUUUGCUUUUU (((-.(((((((((((.((((....)))).......(((((((((((.---...)))))))))))...)))).))))))))))........... ( -29.00) >DroSec_CAF1 210846 83 + 1 GCA-GCCAGCGAUCAAUGUGAUCAAUCGCGCUUCAAUUUUUCUUCUUU---UCAAAGAAGAAAGAGAUUUGGUUUGCUGGUGCUUUU------- (((-.(((((((((((.((((....)))).......(((((((((((.---...)))))))))))...)))).))))))))))....------- ( -29.00) >DroEre_CAF1 226818 79 + 1 GCA-GCCAGCGAUCAAUGUGAUCAAUCGCGCUUCAAUUUUUCUUCUUU---UCAAAGAAGAAAGAGAUUUGGUUUGCUGCUGC----------- (((-((.(((((((((.((((....)))).......(((((((((((.---...)))))))))))...)))).))))))))))----------- ( -27.80) >DroWil_CAF1 246049 90 + 1 GCAAGCAAACGAUCAAUGUGAUCAAUCGUGCUUCAAUUUUUCUUAUGUCUCUCUAACA----CAAGACUUUGUCUUGUAGUAGUAGUAAGUCUU (.(((((...((((.....)))).....)))))).......(((((......(((..(----((((((...)))))))..)))..))))).... ( -19.30) >consensus GCA_GCCAGCGAUCAAUGUGAUCAAUCGCGCUUCAAUUUUUCUUCUUU___UCAAAGAAGAAAGAGAUUUGGUUUGCUGGUGCUUUU_______ (((.(((((((((...((....))))))).......((((((((((((.....))))))))))))....)))).)))................. (-15.96 = -17.52 + 1.56)


| Location | 7,462,542 – 7,462,632 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -14.23 |
| Energy contribution | -15.35 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7462542 90 - 22224390 AAAAAGCAAAAGCACCAGCAAACCAAAUCUCUUUCUUCUUUGA---AAAGAAGAAAAAUUGAAGCGCGAUUGAUCACAUUGAUCGCUGGC-UGC ...........((((((((............(((((((((...---.)))))))))(((((.....)))))((((.....))))))))).-))) ( -24.40) >DroSec_CAF1 210846 83 - 1 -------AAAAGCACCAGCAAACCAAAUCUCUUUCUUCUUUGA---AAAGAAGAAAAAUUGAAGCGCGAUUGAUCACAUUGAUCGCUGGC-UGC -------....((((((((............(((((((((...---.)))))))))(((((.....)))))((((.....))))))))).-))) ( -24.40) >DroEre_CAF1 226818 79 - 1 -----------GCAGCAGCAAACCAAAUCUCUUUCUUCUUUGA---AAAGAAGAAAAAUUGAAGCGCGAUUGAUCACAUUGAUCGCUGGC-UGC -----------...(((((............(((((((((...---.))))))))).........(((((..((...))..)))))..))-))) ( -24.60) >DroWil_CAF1 246049 90 - 1 AAGACUUACUACUACUACAAGACAAAGUCUUG----UGUUAGAGAGACAUAAGAAAAAUUGAAGCACGAUUGAUCACAUUGAUCGUUUGCUUGC ........((.(((.((((((((...))))))----)).))))).................(((((((((..((...))..)))))..)))).. ( -20.80) >consensus _______AAAAGCACCAGCAAACCAAAUCUCUUUCUUCUUUGA___AAAGAAGAAAAAUUGAAGCGCGAUUGAUCACAUUGAUCGCUGGC_UGC .................(((..(((..((..((((((((((.....))))))))))....))...(((((..((...))..))))))))..))) (-14.23 = -15.35 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:46 2006