
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,454,634 – 7,454,782 |
| Length | 148 |
| Max. P | 0.998543 |

| Location | 7,454,634 – 7,454,744 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -28.04 |
| Energy contribution | -27.90 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7454634 110 + 22224390 ----------CCCAAGAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ----------.........((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. ( -28.40) >DroSec_CAF1 203028 110 + 1 ----------CCCAAGAAAGCUCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ----------.........((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...))))))))..........)))).. ( -29.90) >DroEre_CAF1 218451 110 + 1 ----------CCCAAGAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ----------.........((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. ( -28.40) >DroWil_CAF1 234682 110 + 1 ----------ACGGAAAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ----------.........((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. ( -28.40) >DroYak_CAF1 249407 110 + 1 ----------CCCAAGAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ----------.........((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. ( -28.40) >DroAna_CAF1 207364 120 + 1 GGAACCACACCACUGGAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ....(((......)))...((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. ( -30.20) >consensus __________CCCAAGAAAGCCCACGAAUGCAGCCAACAUUGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAA ...................((((..((..((((((..((...(((....(.((((...)))).)...)))...))..))))))))((((((((...)))))))).........)).)).. (-28.04 = -27.90 + -0.14)



| Location | 7,454,634 – 7,454,744 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -32.69 |
| Energy contribution | -32.55 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7454634 110 - 22224390 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUCUUGGG---------- .....(((....((((..(((((((.....)))))(((((((((.((...(((((..((((((....)))))))))))...)).))))))..)))..))..))))..)))---------- ( -33.10) >DroSec_CAF1 203028 110 - 1 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGAGCUUUCUUGGG---------- ..((((.....((((....((((((.....))))))..((((((.((...(((((..((((((....)))))))))))...)).))))))..)))).)))).........---------- ( -33.30) >DroEre_CAF1 218451 110 - 1 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUCUUGGG---------- .....(((....((((..(((((((.....)))))(((((((((.((...(((((..((((((....)))))))))))...)).))))))..)))..))..))))..)))---------- ( -33.10) >DroWil_CAF1 234682 110 - 1 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUUUCCGU---------- ..((.((....((((....((((((.....))))))..((((((.((...(((((..((((((....)))))))))))...)).))))))..)))).)))).........---------- ( -33.00) >DroYak_CAF1 249407 110 - 1 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUCUUGGG---------- .....(((....((((..(((((((.....)))))(((((((((.((...(((((..((((((....)))))))))))...)).))))))..)))..))..))))..)))---------- ( -33.10) >DroAna_CAF1 207364 120 - 1 UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUCCAGUGGUGUGGUUCC (..(.((.....((((..(((((((.....)))))(((((((((.((...(((((..((((((....)))))))))))...)).))))))..)))..))..))))....)).)..).... ( -35.30) >consensus UUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCAAUGUUGGCUGCAUUCGUGGGCUUUCUUGGG__________ ..((((.....((((....((((((.....))))))..((((((.((...(((((..((((((....)))))))))))...)).))))))..)))).))))................... (-32.69 = -32.55 + -0.14)



| Location | 7,454,664 – 7,454,782 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
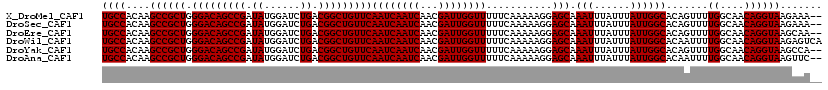
>X_DroMel_CAF1 7454664 118 + 22224390 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAGUUUUGGCAACAGGUAAGAAA-- ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....)))))).....-- ( -32.70) >DroSec_CAF1 203058 118 + 1 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAGUUUUGGCAACAGGUAAGAAA-- ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....)))))).....-- ( -32.70) >DroEre_CAF1 218481 118 + 1 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAGUUUUGGCAACAGGUAAGCAA-- ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....)))))).....-- ( -32.70) >DroWil_CAF1 234712 120 + 1 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAAUUUUGGCAACAGGUAAGAGUCA ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....))))))....... ( -32.70) >DroYak_CAF1 249437 118 + 1 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAGUUUUGGCAACAGGUAAGCCA-- ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....)))))).....-- ( -32.70) >DroAna_CAF1 207404 118 + 1 UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAAUUUUGGCAACAGGUAAGUUC-- ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....)))))).....-- ( -32.70) >consensus UGCCACAAGCCGCUGGGACAGCCGAUAUGGAUCUGACGGCUGUUCAAUCAAUCAACGAUUGGUUUUUCAAAAAGGAGCAAAUUUAUUUAUUGGCACAGUUUUGGCAACAGGUAAGAAA__ ((((....((((((.(((((((((.((......)).)))))))))((((((((...))))))))...........))).(((......)))))).......((....))))))....... (-32.70 = -32.70 + -0.00)


| Location | 7,454,664 – 7,454,782 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
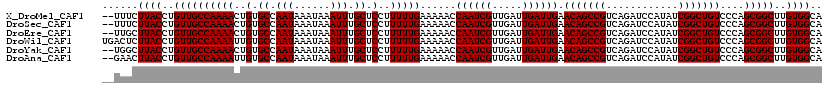
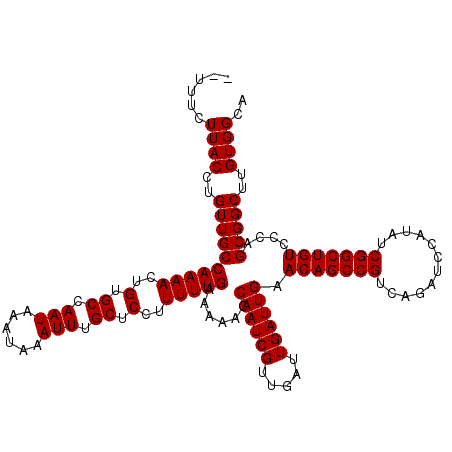
>X_DroMel_CAF1 7454664 118 - 22224390 --UUUCUUACCUGUUGCCAAAACUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA --............(((((.......(((..............................((((((.....)))))).(((((((............)))))))......)))...))))) ( -26.30) >DroSec_CAF1 203058 118 - 1 --UUUCUUACCUGUUGCCAAAACUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA --............(((((.......(((..............................((((((.....)))))).(((((((............)))))))......)))...))))) ( -26.30) >DroEre_CAF1 218481 118 - 1 --UUGCUUACCUGUUGCCAAAACUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA --.((((.((..((((((((((..(.((.(((......))).)).)..)))))......((((((.....)))))).(((((((............)))))))....)))))..)))))) ( -29.10) >DroWil_CAF1 234712 120 - 1 UGACUCUUACCUGUUGCCAAAAUUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA ......((((..((((((((((..(.((.(((......))).)).)..)))))......((((((.....)))))).(((((((............)))))))....)))))..)))).. ( -27.00) >DroYak_CAF1 249437 118 - 1 --UGGCUUACCUGUUGCCAAAACUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA --..(((.((..((((((((((..(.((.(((......))).)).)..)))))......((((((.....)))))).(((((((............)))))))....)))))..))))). ( -29.00) >DroAna_CAF1 207404 118 - 1 --GAACUUACCUGUUGCCAAAAUUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA --....((((..((((((((((..(.((.(((......))).)).)..)))))......((((((.....)))))).(((((((............)))))))....)))))..)))).. ( -27.00) >consensus __UUUCUUACCUGUUGCCAAAACUGUGCCAAUAAAUAAAUUUGCUCCUUUUUGAAAAACCAAUCGUUGAUUGAUUGAACAGCCGUCAGAUCCAUAUCGGCUGUCCCAGCGGCUUGUGGCA ......((((..((((((((((..(.((.(((......))).)).)..)))))......((((((.....)))))).(((((((............)))))))....)))))..)))).. (-26.47 = -26.47 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:42 2006