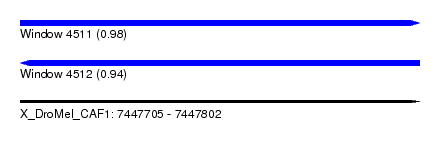
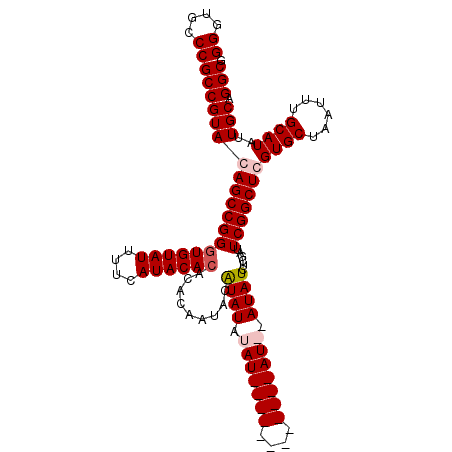
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,447,705 – 7,447,802 |
| Length | 97 |
| Max. P | 0.981725 |

| Location | 7,447,705 – 7,447,802 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -29.10 |
| Energy contribution | -30.97 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7447705 97 + 22224390 CGCCUGCAAUAUGCAAAUUAGCACGAGCCGAAUCGAGAUAU--AU------------AUGUAUAUGUAUUGUAUGUGUAUGAAAAUACACCCGGCUCUACGGCGGGCACCC .((((((....(((......))).((((((......(((((--((------------(....))))))))....((((((....)))))).))))))....)))))).... ( -37.20) >DroSec_CAF1 196039 99 + 1 CGCCUGCAAUAUGCAAAUUAGCACGAGCCGAAUCGAGAUAUGUAU------------AUAUAUAUGUAUUGUGUGUGUAUGAAAAUACACCCGGCUGUACGGCGGGCACCC .((((((....(((......)))..(((((......(((((((((------------....)))))))))....((((((....)))))).))))).....)))))).... ( -34.20) >DroSim_CAF1 190986 99 + 1 CGCCUGCAAUAUGCAAAUUAGCACGAGCCGAAUCGAGAUAUGUAU------------AUAUAUAUGUAUUGUGUGUGUAUGAAAAUACACCCGGCUGUACGGCGGGCACCC .((((((....(((......)))..(((((......(((((((((------------....)))))))))....((((((....)))))).))))).....)))))).... ( -34.20) >DroEre_CAF1 211115 109 + 1 CGCCUGCAAUAUGCAAAUUAGCACGAGCCGAAUCGAGAUAU--AUACAUAUGUACGUAUAUAUACAUACUGUGUGUGUAUGAAAAUACAGCCGGCUCUACGGCGGGCACCC .((((((....(((......))).((((((.........((--(((((((((((........)))))).)))))))((((....))))...))))))....)))))).... ( -39.40) >consensus CGCCUGCAAUAUGCAAAUUAGCACGAGCCGAAUCGAGAUAU__AU____________AUAUAUAUGUAUUGUGUGUGUAUGAAAAUACACCCGGCUCUACGGCGGGCACCC .((((((....(((......))).((((((.............................(((((.....)))))((((((....)))))).))))))....)))))).... (-29.10 = -30.97 + 1.87)

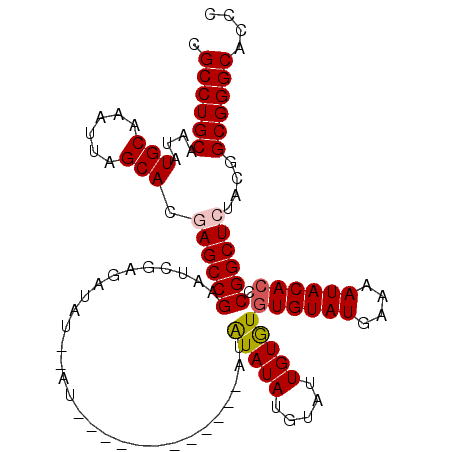
| Location | 7,447,705 – 7,447,802 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 89.49 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.88 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7447705 97 - 22224390 GGGUGCCCGCCGUAGAGCCGGGUGUAUUUUCAUACACAUACAAUACAUAUACAU------------AU--AUAUCUCGAUUCGGCUCGUGCUAAUUUGCAUAUUGCAGGCG ((....))(((((((((((((((((((....)))))).........(((((...------------.)--))))......)))))))((((......))))..))).))). ( -30.20) >DroSec_CAF1 196039 99 - 1 GGGUGCCCGCCGUACAGCCGGGUGUAUUUUCAUACACACACAAUACAUAUAUAU------------AUACAUAUCUCGAUUCGGCUCGUGCUAAUUUGCAUAUUGCAGGCG ((....))(((((((((((((((((((....))))))...........((((..------------....))))......)))))).)))).....(((.....)))))). ( -27.90) >DroSim_CAF1 190986 99 - 1 GGGUGCCCGCCGUACAGCCGGGUGUAUUUUCAUACACACACAAUACAUAUAUAU------------AUACAUAUCUCGAUUCGGCUCGUGCUAAUUUGCAUAUUGCAGGCG ((....))(((((((((((((((((((....))))))...........((((..------------....))))......)))))).)))).....(((.....)))))). ( -27.90) >DroEre_CAF1 211115 109 - 1 GGGUGCCCGCCGUAGAGCCGGCUGUAUUUUCAUACACACACAGUAUGUAUAUAUACGUACAUAUGUAU--AUAUCUCGAUUCGGCUCGUGCUAAUUUGCAUAUUGCAGGCG ((....))(((((((((((((.........(((((.......))))).((((((((((....))))))--))))......)))))))((((......))))..))).))). ( -35.50) >consensus GGGUGCCCGCCGUACAGCCGGGUGUAUUUUCAUACACACACAAUACAUAUAUAU____________AU__AUAUCUCGAUUCGGCUCGUGCUAAUUUGCAUAUUGCAGGCG ((....))(((((((((((((((((((....)))))).........((((.(((((((....))))))).))))......)))))))((((......))))..))).))). (-25.06 = -26.88 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:36 2006