| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,443,233 – 7,443,329 |
| Length | 96 |
| Max. P | 0.983173 |

| Location | 7,443,233 – 7,443,329 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
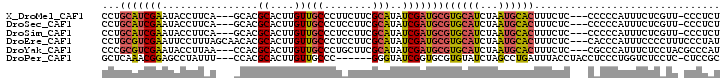
>X_DroMel_CAF1 7443233 96 + 22224390 CCUGCAUCGAAUACCUUCA---GCACGCACUUGUUGCCCUUCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC---CCCCCAUUUCUCGUU-CCCUCU ..(((((.(((....))).---(((((((.(((.(((........)))...)))))))))).....))))).......---...............-...... ( -20.40) >DroSec_CAF1 191306 96 + 1 CCUGCAUCGAAUACCUUCA---GCACGCACUUGUUGCCCUCCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC---CCCCCAUUUCUCGUU-CCCUCU ..(((((.(((....))).---(((((((.(((.(((........)))...)))))))))).....))))).......---...............-...... ( -20.40) >DroSim_CAF1 186565 96 + 1 CCUGCAUCGAAUACCUUCA---GCACGCACUUGUUGCCCUCCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC---CCCCCAUUUCUCGUU-CCCUCU ..(((((.(((....))).---(((((((.(((.(((........)))...)))))))))).....))))).......---...............-...... ( -20.40) >DroEre_CAF1 206564 100 + 1 CCUGCGUCGAAUUCCUUUAGCAACACGCACUUGUUGCCCUCCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC---CACCCAUUUCCCCUUUCCCUAU ...(((((((.........((((((......)))))).............)))))))((((((...))))))......---...................... ( -20.75) >DroYak_CAF1 235983 97 + 1 CCCGCGUCGAAUACCUUAA---CCACGCACUUGUUGCCCUGCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC---CGCCCAUUUCUCCUACGCCCAU ...(((((((.........---....(((.....)))..(((...)))..)))))))((((((...))))))......---...................... ( -18.80) >DroPer_CAF1 275167 93 + 1 GCUCAAACGGAGCCUAUUU---CCACGCACUUGUUGCCC------GGGUAUCGGUGCGUGUAUCUAGCCUGAUUUACCUACCUCCCUGGUCUCCUC-CUCCGC .......(((((.(((..(---.((((((((...(((..------..)))..)))))))).)..)))............(((.....)))......-))))). ( -24.80) >consensus CCUGCAUCGAAUACCUUCA___GCACGCACUUGUUGCCCUCCUUCGCAUAUCGAUGCGUGCAUCUAAUGCACUUUCUC___CCCCCAUUUCUCCUU_CCCUCU ...(((((((................((....))(((........)))..)))))))((((((...))))))............................... (-11.93 = -12.18 + 0.25)
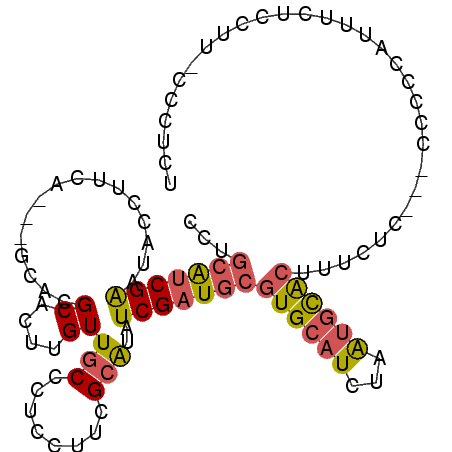
| Location | 7,443,233 – 7,443,329 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -14.20 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7443233 96 - 22224390 AGAGGG-AACGAGAAAUGGGGG---GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGAAGGGCAACAAGUGCGUGC---UGAAGGUAUUCGAUGCAGG ......-..(.(....).)...---......((((((...))))))(((((((.((((..((..(.((.....)).)...)---)....)))))))))))... ( -23.20) >DroSec_CAF1 191306 96 - 1 AGAGGG-AACGAGAAAUGGGGG---GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGGAGGGCAACAAGUGCGUGC---UGAAGGUAUUCGAUGCAGG ......-..(.(....).)...---......((((((...))))))(((((((.((((...((.(.(((.....))).).)---)....)))))))))))... ( -24.30) >DroSim_CAF1 186565 96 - 1 AGAGGG-AACGAGAAAUGGGGG---GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGGAGGGCAACAAGUGCGUGC---UGAAGGUAUUCGAUGCAGG ......-..(.(....).)...---......((((((...))))))(((((((.((((...((.(.(((.....))).).)---)....)))))))))))... ( -24.30) >DroEre_CAF1 206564 100 - 1 AUAGGGAAAGGGGAAAUGGGUG---GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGGAGGGCAACAAGUGCGUGUUGCUAAAGGAAUUCGACGCAGG ...................(((---..(((..(((((..((((....))))...)))))......(((((((......))))))).......)))..)))... ( -24.60) >DroYak_CAF1 235983 97 - 1 AUGGGCGUAGGAGAAAUGGGCG---GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGCAGGGCAACAAGUGCGUGG---UUAAGGUAUUCGACGCGGG ....((((.(((..((((.((.---......)).))))..((((((((.(((.....))).))..(....)..))))))..---........))).))))... ( -26.00) >DroPer_CAF1 275167 93 - 1 GCGGAG-GAGGAGACCAGGGAGGUAGGUAAAUCAGGCUAGAUACACGCACCGAUACCC------GGGCAACAAGUGCGUGG---AAAUAGGCUCCGUUUGAGC ((((((-...((.(((.........)))...))...(((....(((((((((.....)------)(....)..))))))).---...))).))))))...... ( -27.80) >consensus AGAGGG_AACGAGAAAUGGGGG___GAGAAAGUGCAUUAGAUGCACGCAUCGAUAUGCGAAGGAGGGCAACAAGUGCGUGC___UGAAGGUAUUCGAUGCAGG ................................(((((..(((((((((((.....(((........)))....))))))))..........)))..))))).. (-14.20 = -15.03 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:34 2006