
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,433,234 – 7,433,372 |
| Length | 138 |
| Max. P | 0.724704 |

| Location | 7,433,234 – 7,433,337 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.14 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7433234 103 - 22224390 UGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGGCC----------------UAUCUGUAUCCCAUGGAUGCGUGUGU-UUCCCCAUUUUACUUCC (((.((..((((((((((((((..(........)..))))))).)))).))).)).((..----------------(((.(((((((...))))))).))).-..))))).......... ( -34.70) >DroPse_CAF1 229049 92 - 1 UUGA--GUUUGGGUUCCCAUCGUGGUUAACAAUUUGCAAUGGGAAAG----GGGAAGCAC----------------CGGAUGU---UAAUGGCUACAUUUCU-CUC--UGCUUAUCUUUC .(((--((..((((((((((.(..(........)..).)))))))..----((......)----------------)((((((---........))))))))-)..--.)))))...... ( -21.10) >DroSec_CAF1 181746 103 - 1 UGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGGCC----------------UAUCUCUAUCCCAUGGAUGCGUGUGU-UUCCCCAUUUUACUUCC .(((.....((((.((((((((..(........)..))))))))(((((((((((.....----------------..)))))((((...))))..))).))-)..)))).......))) ( -31.90) >DroEre_CAF1 196444 102 - 1 UGGAUCGGCUGGGUUCCCAUCGCGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGCC-----------------UAUCUGUAUCCCAUGGAUGCGUGUGU-UUCCCCAUUUUACUUCC (((.....(((((((((((((((((........)))))))))).)))).))).(((((.-----------------(((..((((((...))))))))).))-))).))).......... ( -36.80) >DroYak_CAF1 224503 120 - 1 UGGAUCGGCUGGGUUCCCAGCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGUGAAAACCUAUGUAUCUGUAUCUGUAUCUGUAUCCCAUGGAUGCGUGUGCCUUCCCCAUUUUAUUUCC (((....(((((....)))))..(((......((..(..(((....)))..)..))....(((((((((((....(((....)))...))))))))))).)))....))).......... ( -34.30) >DroPer_CAF1 262379 92 - 1 UUUA--GUCUGGGUUCCCAUCGCGGUUAACAAUUUGCAAUGGGAAAG----GGGAAGCAC----------------CGGAUGU---UAAUGGCUACAUUUCU-CUC--UGCUUAUCUUUC ....--.......(((((((.((((........)))).)))))))((----(..(((((.----------------.(((.(.---.((((....)))).))-)).--)))))..))).. ( -22.00) >consensus UGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGCCC________________UAUCUGUAUCCCAUGGAUGCGUGUGU_UUCCCCAUUUUACUUCC .........((((((((((((((((........))))))))))))...............................(((.(((((((...))))))).))).....)))).......... (-18.43 = -19.10 + 0.67)

| Location | 7,433,273 – 7,433,372 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -22.19 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.94 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7433273 99 + 22224390 GAUA----------------GGCCUUCUCUGUGGUUUCCCAUCGCAAAUUAUAAGCCACGAUGGGAACCCAGCCGAUCCAUCCAGUCCCAGCCAAUUCAAUCGCAGUACCAAGUA ....----------------(((.....((((((.(((((((((..............))))))))).)))...((....))))).....)))...................... ( -22.44) >DroSec_CAF1 181785 99 + 1 GAUA----------------GGCCUUCUCUGUGGUUUCCCAUCGCAAAUUAUAAGCCACGAUGGGAACCCAGCCGAUCCAUCCAGUCCCAGCCAAUUCAAUCGCAGUACCAAGUA ....----------------(((.....((((((.(((((((((..............))))))))).)))...((....))))).....)))...................... ( -22.44) >DroEre_CAF1 196483 98 + 1 GAUA-----------------GGCUUCUCUGUGGUUUCCCAUCGCAAAUUAUAAGCCGCGAUGGGAACCCAGCCGAUCCAGCCAGUCCCAGCCAAUUCAAUCGCAGUACCAAGUA (((.-----------------((((((.(((.((((.((((((((............)))))))))))))))..))...)))).)))............................ ( -29.80) >DroYak_CAF1 224543 115 + 1 GAUACAGAUACAGAUACAUAGGUUUUCACUGUGGUUUCCCAUCGCAAAUUAUAAGCCACGCUGGGAACCCAGCCGAUCCAUCCAGUCCCAGCCAAUUCAAUCUCACUACCAAACC ....................((((...((((((((((...............)))))))(((((....)))))..........)))...))))...................... ( -20.76) >DroPer_CAF1 262413 86 + 1 UCCG----------------GUGCUUCCC----CUUUCCCAUUGCAAAUUGUUAACCGCGAUGGGAACCCAGAC--UAAAUCCAA-----UCCAAUCCAUUCCCGAUCCCAAA-- ..((----------------(........----..((((((((((............))))))))))....((.--....))...-----............)))........-- ( -15.50) >consensus GAUA________________GGCCUUCUCUGUGGUUUCCCAUCGCAAAUUAUAAGCCACGAUGGGAACCCAGCCGAUCCAUCCAGUCCCAGCCAAUUCAAUCGCAGUACCAAGUA ...............................(((.(((((((((..............))))))))).)))............................................ (-14.30 = -14.94 + 0.64)


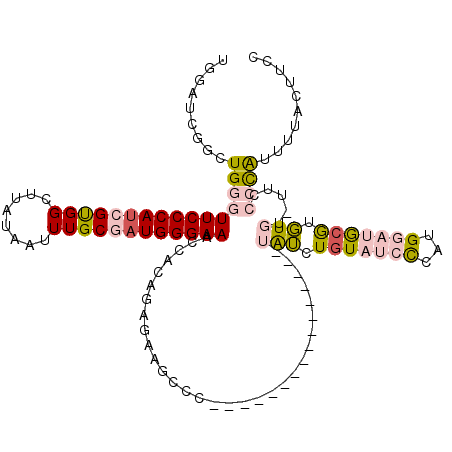
| Location | 7,433,273 – 7,433,372 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -23.46 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7433273 99 - 22224390 UACUUGGUACUGCGAUUGAAUUGGCUGGGACUGGAUGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGGCC----------------UAUC ..((..((....(....).....))..))....(((((..(..((((((((((((((..(........)..))))))).)))).)))....)..)----------------)))) ( -33.00) >DroSec_CAF1 181785 99 - 1 UACUUGGUACUGCGAUUGAAUUGGCUGGGACUGGAUGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGGCC----------------UAUC ..((..((....(....).....))..))....(((((..(..((((((((((((((..(........)..))))))).)))).)))....)..)----------------)))) ( -33.00) >DroEre_CAF1 196483 98 - 1 UACUUGGUACUGCGAUUGAAUUGGCUGGGACUGGCUGGAUCGGCUGGGUUCCCAUCGCGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGCC-----------------UAUC ..((..((....(....).....))..))...((((...((...(((.((((((((((((........)))))))))))).)))..))..))))-----------------.... ( -34.90) >DroYak_CAF1 224543 115 - 1 GGUUUGGUAGUGAGAUUGAAUUGGCUGGGACUGGAUGGAUCGGCUGGGUUCCCAGCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGUGAAAACCUAUGUAUCUGUAUCUGUAUC (((((..((.((..........((((((..(.....)..))))))((.((((((.((..(........)..)).)))))).)).)).)).))))).................... ( -31.20) >DroPer_CAF1 262413 86 - 1 --UUUGGGAUCGGGAAUGGAUUGGA-----UUGGAUUUA--GUCUGGGUUCCCAUCGCGGUUAACAAUUUGCAAUGGGAAAG----GGGAAGCAC----------------CGGA --.......((((....((((((((-----(...)))))--))))...(((((((.((((........)))).)))))))..----........)----------------))). ( -22.90) >consensus UACUUGGUACUGCGAUUGAAUUGGCUGGGACUGGAUGGAUCGGCUGGGUUCCCAUCGUGGCUUAUAAUUUGCGAUGGGAAACCACAGAGAAGCCC________________UAUC ....((((............(..(((((..(.....)..)))))..).((((((((((((........))))))))))))))))............................... (-23.46 = -24.60 + 1.14)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:30 2006