| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,427,099 – 7,427,192 |
| Length | 93 |
| Max. P | 0.592720 |

| Location | 7,427,099 – 7,427,192 |
|---|---|
| Length | 93 |
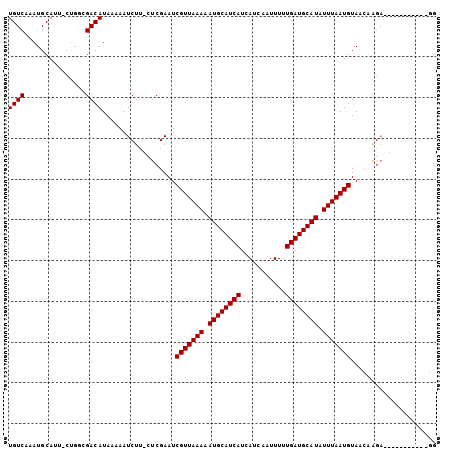
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.09 |
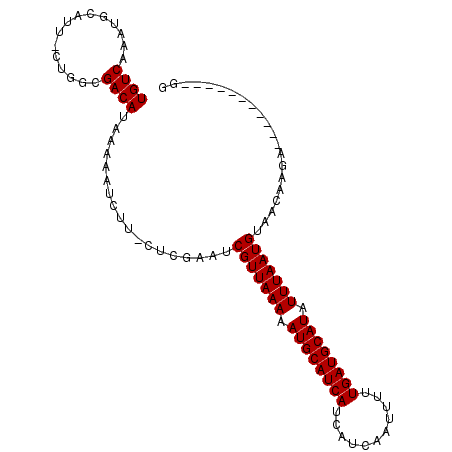
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7427099 93 + 22224390 UGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUC-CUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUAACGAGA-----------GG ((((....((.......)))))).......(((-.(((...(((((((.((((((((...........)))))))).)))))))...)))))-----------). ( -23.70) >DroVir_CAF1 231574 98 + 1 UGUCAAAUGCAU-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUACGUGGACGAGUCAAUGCUG ........((((-------(((..........(((((.(..(((((((.((((((((...........)))))))).)))))))....).)))))))).)))).. ( -25.60) >DroPse_CAF1 221584 93 + 1 UGUCAAAUGCAUUCCUGGCGACAUAAAAAUCUU-CUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUAACAAGA-----------GG ((((....((.......)))))).......(((-((.(...(((((((.((((((((...........)))))))).)))))))...).)))-----------)) ( -20.90) >DroYak_CAF1 217746 93 + 1 UGUCAAAUGCAUUUCUGGCGACAUAAAAAUCUC-CUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUAACUAGA-----------GG ((((....((.......)))))).........(-(((....(((((((.((((((((...........)))))))).)))))))......))-----------)) ( -22.20) >DroMoj_CAF1 266309 84 + 1 UGUCAAAUGCAC-------GACAUAAAAAUCUUUGUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUACGA--------------CG ((((........-------))))...........((((...(((((((.((((((((...........)))))))).)))))))..)))--------------). ( -23.70) >DroPer_CAF1 254901 93 + 1 UGUCAAAUGCAUUCCUGGGGACAUAAAAAUCUU-CUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUAACAAGA-----------GG ...............((....)).......(((-((.(...(((((((.((((((((...........)))))))).)))))))...).)))-----------)) ( -20.90) >consensus UGUCAAAUGCAUU_CUGGCGACAUAAAAAUCUU_CUCGAAUCGUUAAAAAUGCAUCAUCAUCAAUUUUUGAUGCAUAUUUAAUGUAACAAGA___________GG ((((...............))))..................(((((((.((((((((...........)))))))).)))))))..................... (-15.74 = -15.74 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:26 2006