
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,423,080 – 7,423,177 |
| Length | 97 |
| Max. P | 0.877338 |

| Location | 7,423,080 – 7,423,177 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
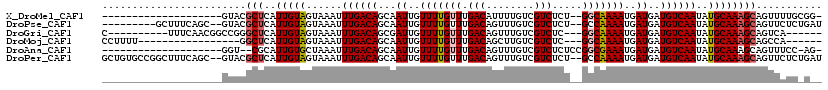
>X_DroMel_CAF1 7423080 97 + 22224390 --------------------GUACGCUCAUUGUAGUAAAUUUGACAGCAAUUGUUUUGUUUGACAUUUUGUCGUCUCU--GGCAAAAUGAUGAUGUCAAUAUGCAAAGCAGUUUUGCGG- --------------------...(((..(((((.(((...((((((...((..(((((((.(((........)))...--)))))))..))..))))))..)))...)))))...))).- ( -27.10) >DroPse_CAF1 216562 107 + 1 ---------GCUUUCAGC--GUACGCUCAUUGUAGUAAAUUUGACAGCAAUUGUUUUGUUUGACAGUUUGUCGUCUCU--GCCAAAAUGAUGAUGUCAAUAUGCAAAGCAGUUCUCUGAU ---------(((.((((.--.(((((.....)).)))...)))).)))(((((((((((((((((.....(((((...--........)))))))))))...)))))))))))....... ( -25.90) >DroGri_CAF1 211370 101 + 1 C----------UUUCAACGGCCGGGCUCAUUGUAGUAAAUUUGACAGCGAUUGUUUUGUUUGACAGUUUGUCGUCUC---GGCAAAAUGAUGAUGUCAAUAUGCAAAGCAGUCA------ (----------(.((((..((...((.....)).))....)))).)).(((((((((((((((((.(((((((...)---)))))).......))))))...))))))))))).------ ( -25.60) >DroMoj_CAF1 260289 94 + 1 CCUUUU-----------------GGCUCAUUGUAGUAAAUUUGACAGCAAUUGUUUUGUUUGACAGCUUGUCGUCUC---GGCAAAAUGAUGAUGUCAAUAUGCAAAGCAGCCA------ .....(-----------------(((((.(((((......((((((...((..(((((((.(((........)))..---)))))))..))..))))))..))))).).)))))------ ( -28.30) >DroAna_CAF1 178855 96 + 1 --------------------GGU--CGCAUUGUGCUAAAUUUGACAGCAAUUGUUUUGUUUGACAGUUUGUCGUCUCUCCGGCGAAAUGAUGAUGUCAAUAUGCAAAGCAGUUUCC-AG- --------------------.((--((.(((......))).))))...((((((((((((((((((((..(((((.....)))))...)))..))))))...)))))))))))...-..- ( -25.50) >DroPer_CAF1 249683 116 + 1 GCUGUGCCGGCUUUCAGC--GUACGCUCAUUGUAGUAAAUUUGACAGCAAUUGUUUUGUUUGACAGUUUGUCGUCUCU--GCCAAAAUGAUGAUGUCAAUAUGCAAAGCAGUUCUCUGAU ((((...))))..((((.--(.(((((..(((((......((((((...((..(((((...(((........)))...--..)))))..))..))))))..)))))))).)).).)))). ( -27.80) >consensus ____________________GUACGCUCAUUGUAGUAAAUUUGACAGCAAUUGUUUUGUUUGACAGUUUGUCGUCUCU__GGCAAAAUGAUGAUGUCAAUAUGCAAAGCAGUUAUC_G__ ........................(((..(((((......((((((...((..(((((((.(((........))).....)))))))..))..))))))..))))))))........... (-19.53 = -19.92 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:25 2006