| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,418,651 – 7,418,748 |
| Length | 97 |
| Max. P | 0.970831 |

| Location | 7,418,651 – 7,418,748 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.53 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.63 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7418651 97 + 22224390 AGCAA-----------------A-CAAA---C-AAACA-AACAACAAAAGUCAAAUAGAGCGAGGCAAGAGCGGCACAAAAAUUGCUGUUCGAAAAUUGUUGCGCUUUGGGUUUGCGUGU .((((-----------------(-(...---.-.....-....((....))....(((((((..((((((((((((.......)))))))).....))))..))))))).)))))).... ( -28.00) >DroPse_CAF1 211733 110 + 1 ACCAC-A---UCGG-CAGACAACAGCAA---C-AAAAGCUACAGCAAAAGUCAAAUAGAGCGAAGCA-GAGCGGCACAAAAAUUGCAGUUCGAAAAUUGUUGCGCUUUCGGUUUGCGUGU ...((-(---(..(-(((((....((((---(-((..(((...((..............))..))).-((((.(((.......))).)))).....))))))).......)))))))))) ( -27.84) >DroSec_CAF1 167519 97 + 1 AGCAA-----------------A-CAAA---C-AAACA-AACAACAAAAGUCAAAUAGAGCGAGGCAAGAGCGGCACAAAAAUUGCUGUUCAAAAAUUGUUGCGCUUUGGGUUUGCGUGU .((((-----------------(-(...---.-.....-....((....))....(((((((..((((((((((((.......)))))))).....))))..))))))).)))))).... ( -28.00) >DroYak_CAF1 207188 116 + 1 AGGCAACAAACCAACCAAACAAACCAAA---C-AAACAAAACAACAAAAGUCAAAUAGAGCGCGGCAAGAGCGGCACACAAAUUGCUGUUCGAAAAUUGUUGCGCUUUGGGUUUGCGUGU .(....)........((..((((((...---.-..........((....))....(((((((((((((((((((((.......)))))))).....)))))))))))))))))))..)). ( -34.60) >DroAna_CAF1 174169 112 + 1 ACCAC-AACACAA-----ACACAGCACAGCACAGAACA-ACAAAAAAAAGUCAAAUAGAGCGAGGCA-GAGCGGCACAAAAAUUGCAGUUCGAAAAUUGUUGCGCUUCGGGUUUGCGUGU .....-.((.(((-----((..(((.(((((.......-..........(((...........))).-((((.(((.......))).))))......))))).)))....))))).)).. ( -23.00) >DroPer_CAF1 244813 110 + 1 ACCAC-A---UCGG-CACACAACAGCAA---C-AAAAGCUACAGCAAAAGUCAAAUAGAGCGAAGCA-GAGCGGCACAAAAAUUGCAGUUCGAAAAUUGUUGCGCUUUCGGUUUGCGUGU .....-.---...(-(((......((((---(-((..(((...((..............))..))).-((((.(((.......))).)))).....)))))))((.........)))))) ( -24.94) >consensus ACCAA_A____C______ACAAA_CAAA___C_AAACA_AACAACAAAAGUCAAAUAGAGCGAGGCA_GAGCGGCACAAAAAUUGCAGUUCGAAAAUUGUUGCGCUUUGGGUUUGCGUGU ...........................................(((...(.(((((.(((((.((((.((((.(((.......))).))))......)))).)))))...)))))).))) (-17.91 = -17.63 + -0.28)

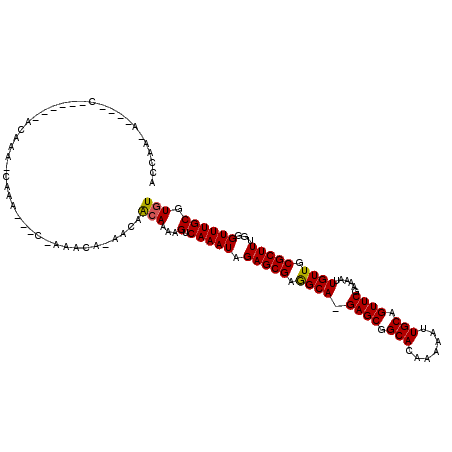

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:21 2006