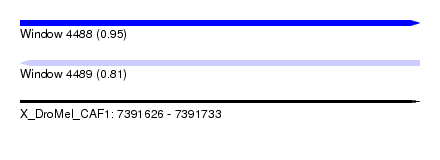
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,391,626 – 7,391,733 |
| Length | 107 |
| Max. P | 0.950103 |

| Location | 7,391,626 – 7,391,733 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.35 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
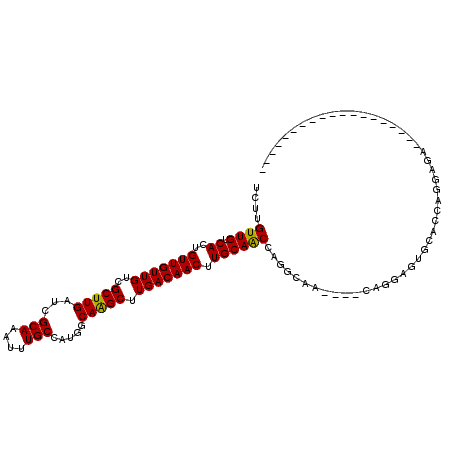
>X_DroMel_CAF1 7391626 107 + 22224390 UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAGCAAACCAA----CAGAAAUACACCUUGAGGAAAAAGUCACAAAAUUA-- ..((((((...((((.(((((((((((...(((....))).....)))))..))))))..)))).....)))----))).......((....)).................-- ( -24.20) >DroVir_CAF1 187161 94 + 1 CCAUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAACCAGGCAAGCGGCAGCGGUGCAGAGGCAGA------------------- .....(((((.((((((((((((((((...((((.(((((....))))).))).)..((((....))))))))))))))))..)))).))))).------------------- ( -35.90) >DroPse_CAF1 179845 86 + 1 UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAACUUUGCAA----CGGGAG-GCUCCACG---------------------- ((((((((.((..((((((((((((((...(((....))).....)))))..))))....)))))..)))))----))))).-........---------------------- ( -26.20) >DroYak_CAF1 177189 109 + 1 UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAACGAGCCAA----CAAGGAUACACACGGAGAAAUAAAUCAAACAAAUACA ((((((((((..(((.(((((((((((...(((....))).....)))))..))))))..)))..))..)))----)))))................................ ( -25.10) >DroAna_CAF1 150170 102 + 1 UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAGGCUUGACAACUUGCAGCCAGGCUG----CCAAAGGGCACUGGGAGAAAAGCAGGGG-GA------ (((((((.((.((((.(((((((((((...(((....))).....)))))..))))))..))))((((..((----((....))))))))..))..))))))).-..------ ( -35.30) >DroPer_CAF1 212690 86 + 1 UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAACUUUGCAA----CGGGAG-GCUCCAUG---------------------- ((((((((.((..((((((((((((((...(((....))).....)))))..))))....)))))..)))))----))))).-........---------------------- ( -26.20) >consensus UCUUGUUGUCACUGUUGUUGUCGCUUGAUCGCAAAUUUGCCAUGGCAAGCUUGACAACUUGCAACCAGGCAA____CAGGAGUGCACCAGGAGA___________________ ....((((.((..(((((((..(((((...(((....))).....))))).))))))).))))))................................................ (-18.74 = -18.38 + -0.36)

| Location | 7,391,626 – 7,391,733 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.35 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.08 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
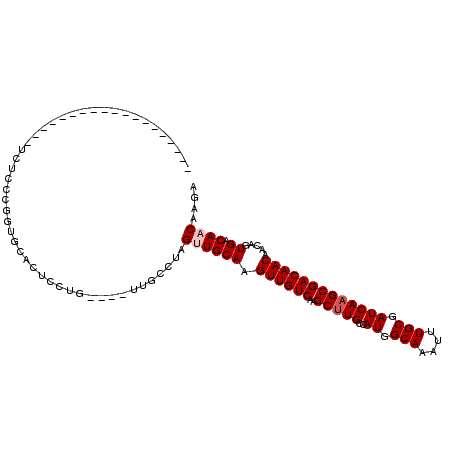
>X_DroMel_CAF1 7391626 107 - 22224390 --UAAUUUUGUGACUUUUUCCUCAAGGUGUAUUUCUG----UUGGUUUGCUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA --...((((((.(((((......)))))((....(((----(((((((((((((((((....)))))))...)))))))((((......))))...))))))..)).)))))) ( -30.10) >DroVir_CAF1 187161 94 - 1 -------------------UCUGCCUCUGCACCGCUGCCGCUUGCCUGGUUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAUGG -------------------..(((....))).(((((....((((.((((((((((((((((.(.....).)))).)))))))))))).))))......)))))......... ( -26.50) >DroPse_CAF1 179845 86 - 1 ----------------------CGUGGAGC-CUCCCG----UUGCAAAGUUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA ----------------------((.((...-..))))----(((((....)))))(((((((.(((((..((.(((....))).)))))))...........))))))).... ( -23.10) >DroYak_CAF1 177189 109 - 1 UGUAUUUGUUUGAUUUAUUUCUCCGUGUGUAUCCUUG----UUGGCUCGUUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA ...........(((.(((........))).)))((((----(((((.....))..((((((..(((((..((.(((....))).)))))))))))))........))))))). ( -27.00) >DroAna_CAF1 150170 102 - 1 ------UC-CCCCUGCUUUUCUCCCAGUGCCCUUUGG----CAGCCUGGCUGCAAGUUGUCAAGCCUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA ------..-....(((.......((((((((....))----))..))))..))).(((((((.(((......)))....((((......)))).........))))))).... ( -27.90) >DroPer_CAF1 212690 86 - 1 ----------------------CAUGGAGC-CUCCCG----UUGCAAAGUUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA ----------------------...((...-...))(----(((((..(((....((((((..(((((..((.(((....))).))))))))))))))))..)).)))).... ( -22.70) >consensus ___________________UCUCCCGGUGCACUCCUG____UUGCCUAGUUGCAAGUUGUCAAGCUUGCCAUGGCAAAUUUGCGAUCAAGCGACAACAACAGUGACAACAAGA ................................................((((((.((((((..(((((..((.(((....))).))))))))))))).....)).)))).... (-17.58 = -18.08 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:16 2006