
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,363,552 – 7,363,697 |
| Length | 145 |
| Max. P | 0.819302 |

| Location | 7,363,552 – 7,363,667 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -11.46 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7363552 115 + 22224390 AUAAAUAGUUUAGGUAAAAGCCUUCGUUCAAAUAUACAAACAAGAUUUUUUCGCAGUCU---GCCCAAGGGCAAGGAUAAUAUUCGUAGCCCAAGCAGACGGCGAAAAAG--GAAAAUUC .......((((((((....))))..((........))))))....(((((((((.((((---((....((((.(.((......)).).))))..)))))).)))))))))--........ ( -33.60) >DroSec_CAF1 112303 113 + 1 AUAAAUAGUUUGCUUUUA--CAUUCGUUCAAAUAUGCAACCAAGAUUUUUCCGCAGUCU---GCCCAAGGGCAAGGAUAAUACUCGUAGCGCAAGCAGACACCGAAAAAG--GAAAACUU ......(((((.(((((.--..((((........(((...............)))((((---(((....)))................((....))))))..))))))))--).))))). ( -22.86) >DroSim_CAF1 112199 113 + 1 AUAAAUAGUUUGCUUUUA--CCUUCGUUCAAAUAUGCAACCAAGAUUUUUCCGCAGUCU---GCCCAAGGGCAAUGAUAAUACUCGUAGCGCAAGCAGACACCGAAAAAG--GAAAACUC .......((((((((...--.(((.(((((....)).))).))).......(((....(---(((....))))((((......)))).))).)))))))).((......)--)....... ( -24.20) >DroEre_CAF1 123687 103 + 1 AUAAAUAGUUU---------CCGUCCUUCAAAUAUAAAACAAAGAUUUUUCCGCAGUCUGCUGCCCAAGCGCAUGGAAAAU------AGCCCAGGCAGACGCCAAAUGGA--GGAAAUCC .......((((---------((.(((.................((....)).((.(((((((((......)).(((.....------...)))))))))))).....)))--)))))).. ( -24.10) >DroYak_CAF1 145986 105 + 1 AUGAAUAGUUA---------CCUUCCCUUGAACAUGAAACAAAGAUUUUUCCGCAGUCUGCUGCCCAAGGGCAAGGAUAAU------AGCCCAAGCAGACGCCAAAAGGAGUGGAAAUCC ...........---------...........................(((((((.((((((((((....)))..((.....------...)).))))))).((....)).)))))))... ( -28.40) >consensus AUAAAUAGUUU___U__A__CCUUCGUUCAAAUAUGCAACCAAGAUUUUUCCGCAGUCU___GCCCAAGGGCAAGGAUAAUA_UCGUAGCCCAAGCAGACGCCGAAAAAG__GAAAAUUC .............................................((((((.((.((((...(((....)))................((....)))))))).))))))........... (-11.46 = -12.30 + 0.84)

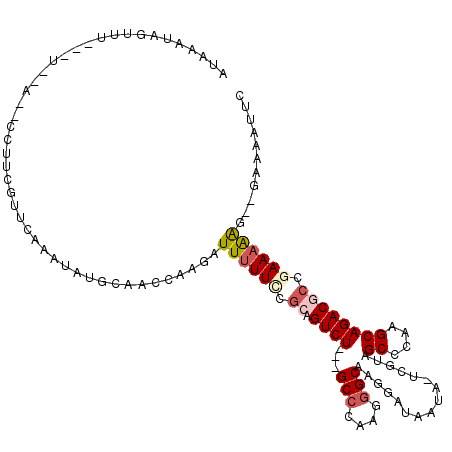
| Location | 7,363,592 – 7,363,697 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.50 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7363592 105 + 22224390 CAAGAUUUUUUCGCAGUCU---GCCCAAGGGCAAGGAUAAUAUUCGUAGCCCAAGCAGACGGCGAAAAAG--GAAAAUUCAAGGGAAGAU----------UGGUUUGAAUCGAUGGCGAC .....(((((((((.((((---((....((((.(.((......)).).))))..)))))).)))))))))--................((----------((((....))))))...... ( -31.80) >DroSec_CAF1 112341 105 + 1 CAAGAUUUUUCCGCAGUCU---GCCCAAGGGCAAGGAUAAUACUCGUAGCGCAAGCAGACACCGAAAAAG--GAAAACUUCAGGGAAGAU----------UGGUUUGGAUCGAUGGCGAC ...(((((((((...((((---(((....)))................((....)))))).((......)--).........))))))))----------)........(((....))). ( -26.70) >DroSim_CAF1 112237 105 + 1 CAAGAUUUUUCCGCAGUCU---GCCCAAGGGCAAUGAUAAUACUCGUAGCGCAAGCAGACACCGAAAAAG--GAAAACUCCAGGGAAGAU----------UGGUUUCGAUCGAUGGCGAC ...(((((((((...((((---(((....))).((((......)))).((....)))))).........(--((....))).))))))))----------)........(((....))). ( -27.70) >DroEre_CAF1 123718 102 + 1 AAAGAUUUUUCCGCAGUCUGCUGCCCAAGCGCAUGGAAAAU------AGCCCAGGCAGACGCCAAAUGGA--GGAAAUCCAAGGGAAGAU----------GGGCUUGGAUCGAUGGCGAC ............((((....))))....((.(((.((....------(((((((((....)))...((((--.....))))........)----------)))))....)).)))))... ( -32.30) >DroYak_CAF1 146017 114 + 1 AAAGAUUUUUCCGCAGUCUGCUGCCCAAGGGCAAGGAUAAU------AGCCCAAGCAGACGCCAAAAGGAGUGGAAAUCCAAGGGGAGAAGAGGGGAGCAGGGUGCGGAUCGAUGGCGAC ....(((..((((((.((((((.(((..((((.........------.)))).......(.((....(((.......)))....)).)....))).)))))).))))))..)))...... ( -43.00) >consensus CAAGAUUUUUCCGCAGUCU___GCCCAAGGGCAAGGAUAAUA_UCGUAGCCCAAGCAGACGCCGAAAAAG__GAAAAUUCAAGGGAAGAU__________UGGUUUGGAUCGAUGGCGAC ....((((((((...((((...(((....)))................((....))))))............((....))..))))))))...................(((....))). (-18.06 = -17.50 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:09 2006