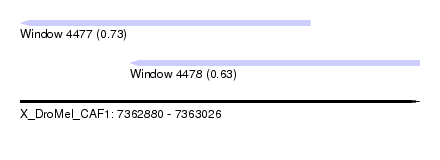
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,362,880 – 7,363,026 |
| Length | 146 |
| Max. P | 0.732121 |

| Location | 7,362,880 – 7,362,986 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.74 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
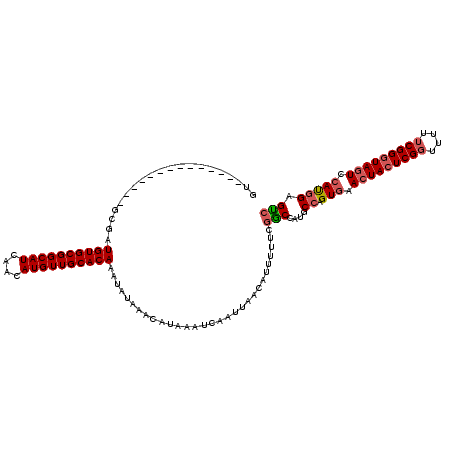
>X_DroMel_CAF1 7362880 106 - 22224390 GU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAUGCCAGUGAACUACUCGGUUUUUCGGGUAGUCAACGGAGCC ..--------------((..((((((((((....))))))))))...........................((((((...))).((..(((((((((....)))))))))..))))))). ( -31.30) >DroSec_CAF1 111624 106 - 1 GU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAUACCAGUGAACUACUCGGUUUUUCGGGUAGUCCAUGGAGUC ..--------------....((((((((((....)))))))))).....................................(((.((.(((((((((....))))))))).))))).... ( -28.60) >DroSim_CAF1 111520 106 - 1 GU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAUACCAGUGAACUACUCGGUUUUUCGGGUAGUCCAUGGAGUC ..--------------....((((((((((....)))))))))).....................................(((.((.(((((((((....))))))))).))))).... ( -28.60) >DroEre_CAF1 123039 119 - 1 G-GCGGUGGCGGCGGCGCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGACCAUGCCAGUGAACUACUCGGUUUCGCGGGUAGUCCACGGAGUC (-(((.(((((((...))..((((((((((....)))))))))).............................)).))))))).(((.((((((((......)))))))).)))...... ( -38.40) >DroYak_CAF1 145290 113 - 1 GUGCGGU-------GCGCGUUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGACCAUGCCAGUGAACUACUCGGUUUUUCGGCUACUCCAUCGAGUG ((((...-------)))).(((((((((((....)))))))))))..........................(((((....(((...(((((....)))))...))).......))))).. ( -26.50) >consensus GU______________GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAUGCCAGUGAACUACUCGGUUUUUCGGGUAGUCCAUGGAGUC ....................((((((((((....))))))))))..............................(((....((.(((.(((((((((....))))))))).))))).))) (-24.82 = -25.38 + 0.56)

| Location | 7,362,920 – 7,363,026 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.73 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -26.36 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
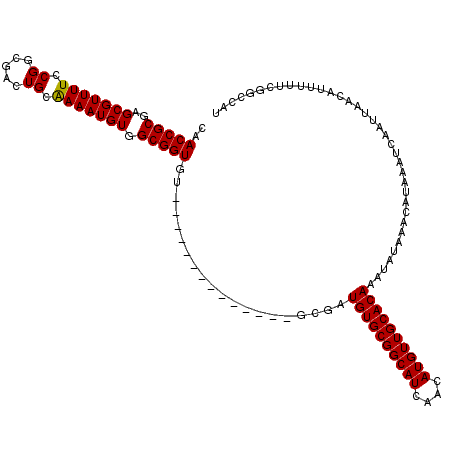
>X_DroMel_CAF1 7362920 106 - 22224390 CAACCGCGAGCGUUUUCCGGCGACUGCGAAAUGUGGCGGUGU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAU ..(((((..(((((((.(((...))).))))))).)))))..--------------....((((((((((....)))))))))).................................... ( -27.80) >DroSec_CAF1 111664 106 - 1 CAACCGCGAGCGUUUUCCGGCGACUGCAAAAUGUGGCGGUGU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAU ..(((((..(((((((.(((...))).))))))).)))))..--------------....((((((((((....)))))))))).................................... ( -27.40) >DroSim_CAF1 111560 106 - 1 CAACCGCGAGCGUUUUCCGGCGACUGCAAAAUGUGGCGGUGU--------------GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAU ..(((((..(((((((.(((...))).))))))).)))))..--------------....((((((((((....)))))))))).................................... ( -27.40) >DroEre_CAF1 123079 119 - 1 CAACCGCGAGCGUUUUCCGCCGACUGCAAAAUGUGGCGGUG-GCGGUGGCGGCGGCGCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGACCAU ....((((.(((((..(((((.(((((........))))))-)))).))).))..)))).((((((((((....)))))))))).................................... ( -40.30) >DroYak_CAF1 145330 113 - 1 CAACCGCGAGCGUUUUCCGCCGACUGCAAAAUGUGGCGGUGUGCGGU-------GCGCGUUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGACCAU ......(((((((...((((..(((((........)))))..)))).-------)))).(((((((((((....)))))))))))...........................)))..... ( -35.50) >consensus CAACCGCGAGCGUUUUCCGGCGACUGCAAAAUGUGGCGGUGU______________GCGAUGUGCGGCAUCAACAUGUUGCACAAAUAUAAACAUAAAUCAAUUAACAUUUUUCGGCCAU ..(((((..(((((((.((.....)).))))))).)))))....................((((((((((....)))))))))).................................... (-26.36 = -26.20 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:07 2006