
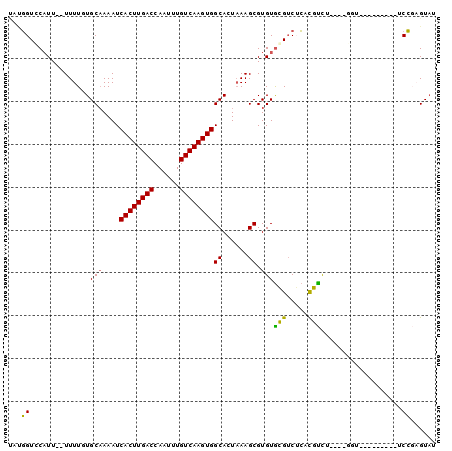
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,315,741 – 7,315,831 |
| Length | 90 |
| Max. P | 0.610237 |

| Location | 7,315,741 – 7,315,831 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -13.76 |
| Energy contribution | -12.65 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
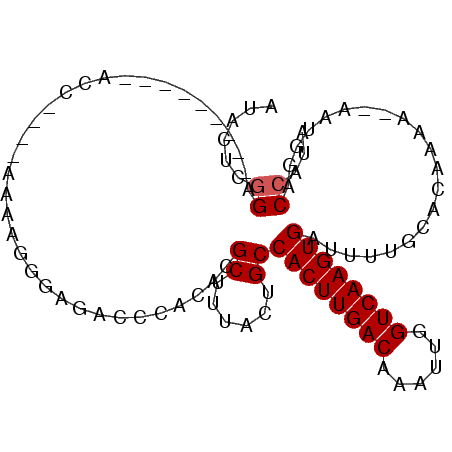
>X_DroMel_CAF1 7315741 90 + 22224390 UUUGGUCCAUU--UUUUGUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCAGUAAAGCGUGUGGGUCUCACUUUUUCCGGGUA--------UCCGAGUAU ...(..((((.--((((((((.....((((((((......))))))))))).)))))...))))..)..........(((...--------.)))..... ( -23.30) >DroPse_CAF1 98155 80 + 1 CAUGGUCCAUU--UUUUCUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCACUAAAGCGUGUGCGUCGCCCGCC-----------------UCCUCGUA- ...(((.....--.............((((((((......))))))))((((........)))).......)))-----------------........- ( -18.90) >DroEre_CAF1 76190 91 + 1 U-CGGUCCAUU--UUUUGUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCAGUAAAGCGUGUGGGGCUCACUUUUUCCGGGUAUU------UCCGAGUAU (-((((((((.--((((((((.....((((((((......))))))))))).)))))...)))))((((.........))))...------.)))).... ( -23.60) >DroYak_CAF1 93592 98 + 1 UACGGUCCAUU--UUUUGUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCAGUAAAGCGUGUGGGUCUCACUUUUUCCGGGUUUUCGGCUAUCCGAGUAU ((((..((((.--((((((((.....((((((((......))))))))))).)))))...))))..)..........(((((........))))).))). ( -27.00) >DroMoj_CAF1 114246 88 + 1 A--UGUGCCUUGCUUUUUUGCCAAAUCACUUGACCAAUUUGUCAAGUGGCACUUAAGCGUGUACGUCUGCCGUCG----CAUCCC------UCGGAGUCC .--.((((..(((((...((((.....(((((((......)))))))))))...))))).))))(.((.(((..(----....).------.))))).). ( -25.40) >DroPer_CAF1 128303 80 + 1 CAUGGUCCAUU--UUUUCUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCACUAAAGCGUGUGCGUCGCCUGCC-----------------UCCUCGUA- ...(((.....--.............((((((((......))))))))((((........))))...)))....-----------------........- ( -18.90) >consensus UAUGGUCCAUU__UUUUGUGCAAAAUCACUUGACCAAUUUGUCAAGUGGCACUAAAGCGUGUGCGUCUCACGUCU____GGU_________UCCGAGUAU ...((..............(((....((((((((......))))))))((......)).)))(((.....)))...................))...... (-13.76 = -12.65 + -1.11)


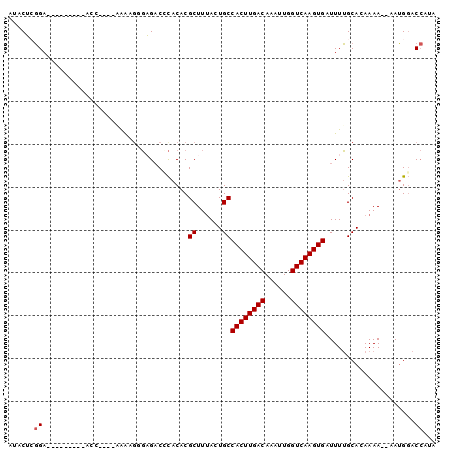
| Location | 7,315,741 – 7,315,831 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -21.37 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7315741 90 - 22224390 AUACUCGGA--------UACCCGGAAAAAGUGAGACCCACACGCUUUACUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCACAAAA--AAUGGACCAAA ....((((.--------...)))).....(..(((.......((......))((((((((......)))))))).)))..)......--........... ( -19.40) >DroPse_CAF1 98155 80 - 1 -UACGAGGA-----------------GGCGGGCGACGCACACGCUUUAGUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCAGAAAA--AAUGGACCAUG -(((...((-----------------((((.((...))...)))))).))).((((((((......))))))))(((((......))--)))........ ( -20.80) >DroEre_CAF1 76190 91 - 1 AUACUCGGA------AAUACCCGGAAAAAGUGAGCCCCACACGCUUUACUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCACAAAA--AAUGGACCG-A ....((((.------.......(.(((((((((((.......)).)))))..((((((((......)))))))).)))).).((...--..))..)))-) ( -20.20) >DroYak_CAF1 93592 98 - 1 AUACUCGGAUAGCCGAAAACCCGGAAAAAGUGAGACCCACACGCUUUACUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCACAAAA--AAUGGACCGUA ....((((....)))).....(((..((((((.(.....).))))))..(((((((((((......)))))))).....))).....--......))).. ( -22.80) >DroMoj_CAF1 114246 88 - 1 GGACUCCGA------GGGAUG----CGACGGCAGACGUACACGCUUAAGUGCCACUUGACAAAUUGGUCAAGUGAUUUGGCAAAAAAGCAAGGCACA--U .......((------((..((----((.(....).))))..).)))..((((((((((((......)))))))......((......))..))))).--. ( -25.00) >DroPer_CAF1 128303 80 - 1 -UACGAGGA-----------------GGCAGGCGACGCACACGCUUUAGUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCAGAAAA--AAUGGACCAUG -........-----------------.(((((....((((........))))((((((((......))))))))..)))))......--........... ( -20.00) >consensus AUACUCGGA_________ACC____AAAAGGGAGACCCACACGCUUUACUGCCACUUGACAAAUUGGUCAAGUGAUUUUGCACAAAA__AAUGGACCAUA ......((..................................((......))((((((((......)))))))).....................))... (-13.63 = -13.80 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:55 2006