
| Sequence ID | X_DroMel_CAF1 |
|---|---|
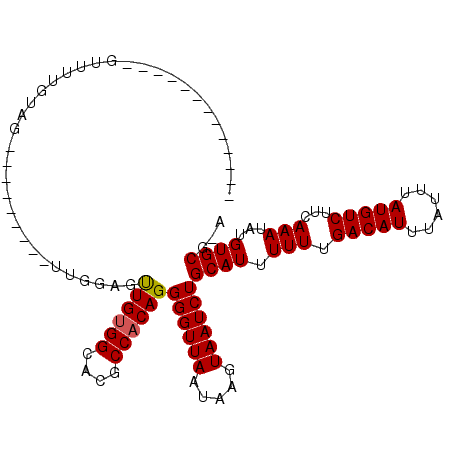
| Location | 7,301,778 – 7,301,874 |
| Length | 96 |
| Max. P | 0.936707 |

| Location | 7,301,778 – 7,301,874 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7301778 96 + 22224390 -------------GUUUUUUCAUUUUUUUU--UUUUUUUGUGGCGCGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCGGA -------------........((((...((--..(((((((((....)))))))))..)).))))..(((((((.(((.(((((......)))))...)))...))))))) ( -21.60) >DroVir_CAF1 71868 88 + 1 -------------GAGCUGGAG---------CUGGAGCUGGGGCACGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCC-G -------------.((((....---------....))))..((((((.....((((...((((((((....((.....))....))))))))..)))).....))))))-. ( -21.00) >DroGri_CAF1 67811 88 + 1 -------------GUGUUGGAG---------UUGGAGUUGUGGCACGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCG-A -------------(..(.....---------((((((((((((....))))))......((((((((....((.....))....))))))))..))))))....)..).-. ( -18.70) >DroSim_CAF1 50667 97 + 1 -------------GUUUUUUCAUUUUUUUUUUUUUUUUUGUGGCGCGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCG-A -------------........................((((((....))))))((((((.....))))))((((.(((.(((((......)))))...)))...)))).-. ( -18.30) >DroYak_CAF1 78365 86 + 1 -------------GUUUUUU-----------UUUUUUUUGUGGCGCGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCG-U -------------.......-----------......((((((....))))))((((((.....))))))((((.(((.(((((......)))))...)))...)))).-. ( -18.30) >DroMoj_CAF1 86223 101 + 1 GGAGACGGAACAGGAACUGAAG---------CUGGAGUUGUGGCACGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCG-G .....((..(((.....(((((---------(.....((((((....))))))((((((.....)))))))).......(((((......)))))))))....))).))-. ( -21.30) >consensus _____________GUUUUGUAG_________UUGGAGUUGUGGCACGCCACAGGGGUUAAUAAGUAAUCUGCAUUUUUUGACAUUUAUUUAUGUCUUCAAAUAUGUGCG_A .....................................((((((....))))))((((((.....))))))((((.(((.(((((......)))))...)))...))))... (-17.82 = -17.85 + 0.03)

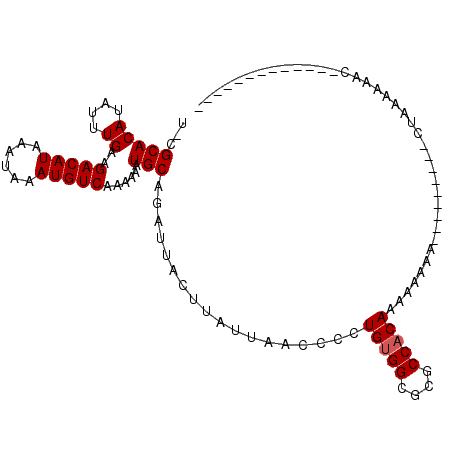
| Location | 7,301,778 – 7,301,874 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.23 |
| Mean single sequence MFE | -14.91 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7301778 96 - 22224390 UCCGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGCGCCACAAAAAAA--AAAAAAAAUGAAAAAAC------------- ((.(((((....))..(((((......)))))......))).))...............(((((....)))))......--.................------------- ( -14.20) >DroVir_CAF1 71868 88 - 1 C-GGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGUGCCCCAGCUCCAG---------CUCCAGCUC------------- .-(((((.........(((((......))))).......((((...............))))...)))))..((((....---------....)))).------------- ( -17.96) >DroGri_CAF1 67811 88 - 1 U-CGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGUGCCACAACUCCAA---------CUCCAACAC------------- .-.(((((....))..(((((......)))))......)))..................(((((....))))).......---------.........------------- ( -14.10) >DroSim_CAF1 50667 97 - 1 U-CGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGCGCCACAAAAAAAAAAAAAAAAAUGAAAAAAC------------- .-.(((((....))..(((((......)))))......)))..................(((((....))))).........................------------- ( -14.10) >DroYak_CAF1 78365 86 - 1 A-CGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGCGCCACAAAAAAAA-----------AAAAAAC------------- .-.(((((....))..(((((......)))))......)))..................(((((....))))).......-----------.......------------- ( -14.10) >DroMoj_CAF1 86223 101 - 1 C-CGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGUGCCACAACUCCAG---------CUUCAGUUCCUGUUCCGUCUCC .-((.(((...((((((((((......)))))...........................(((((....))))).......---------.)))))....)))..))..... ( -15.00) >consensus U_CGCACAUAUUUGAAGACAUAAAUAAAUGUCAAAAAAUGCAGAUUACUUAUUAACCCCUGUGGCGCGCCACAAAAAAAA_________CUAAAAAAC_____________ ...(((((....))..(((((......)))))......)))..................(((((....)))))...................................... (-13.17 = -13.33 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:48 2006