| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,300,492 – 7,300,584 |
| Length | 92 |
| Max. P | 0.701313 |

| Location | 7,300,492 – 7,300,584 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -14.36 |
| Energy contribution | -13.97 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
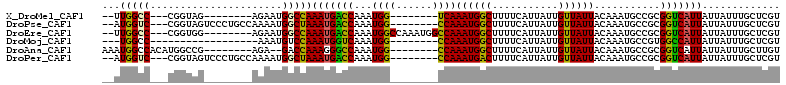
>X_DroMel_CAF1 7300492 92 + 22224390 --UUGGCC---CGGUAG--------AGAAUGGCCAAAUGACCAAAUGG--------UCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUCGU --.(((((---(((((.--------.(((.(((((..(((((....))--------)))..))))).)))....((((....)))).))))).)))))............... ( -33.20) >DroPse_CAF1 81742 100 + 1 --AUGGUC---CGGUAGUCCCUGCCAAAAUGGCUAAAUGACCAAAUGG--------CCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUCGU --(((..(---(((((......((((...((((((..........)))--------)))..)))).........((((....)))).))))).)..))).............. ( -24.60) >DroEre_CAF1 60690 100 + 1 --UUGGCC---CGGUGG--------AGAAUGGCCAAAUGACCAAAUGGCCAAAUGGCCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUCGU --.(((((---(((((.--------..((((((..((((((((..(((((....)))))..)))....)))))...)))))).....))))).)))))............... ( -30.90) >DroMoj_CAF1 83458 84 + 1 ---UGGCC------------------AAAUGUCCAAAUGGUCAAAUGG--------CCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGUGGCCAUUAUUAUUUGCUCGU ---(((((------------------(..((.((....)).))..)))--------)))(((((((...((((..(((....)))))))....)))))))............. ( -24.50) >DroAna_CAF1 53676 95 + 1 AAAUGGCCACAUGGCCG--------AGA--GACCAAAGGGCCAAAUGG--------CCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUUGU .(((((((...((((.(--------(((--..(((...((((....))--------))...)))..))))....((((....))))..)))).)))))))............. ( -29.70) >DroPer_CAF1 110731 100 + 1 --AUGGUC---CGGUAGUCCCUGCCAAAAUGGCUAAAUGACCAAAUGG--------CCAAAUGACUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUCGU --.(((((---.((....))..(((.....))).....)))))(((((--------((.(((((....))))).((((....)))).......)))))))............. ( -23.10) >consensus __AUGGCC___CGGUAG________AAAAUGGCCAAAUGACCAAAUGG________CCAAAUGGCUUUUCAUUAUUGUUAUUACAAAUGCCGCGGUCAUUAUUAUUUGCUCGU ...(((((......................)))))(((((((...((((......))))((((((...........))))))...........)))))))............. (-14.36 = -13.97 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:47 2006