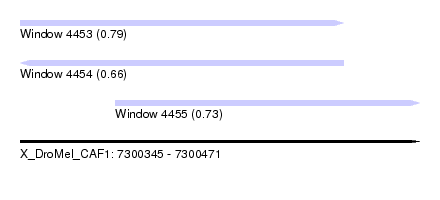
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,300,345 – 7,300,471 |
| Length | 126 |
| Max. P | 0.790917 |

| Location | 7,300,345 – 7,300,447 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -15.96 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7300345 102 + 22224390 UGUGUUUUUUUU--AACCGUGUAUUUUC--GC----UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU-------- ...........(--(((.(((......)--))----.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))))))))).)))).-------- ( -21.80) >DroSec_CAF1 50255 102 + 1 UGUGUUUUUUUU--AACCGUGUAUUUUC--GC----UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU-------- ...........(--(((.(((......)--))----.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))))))))).)))).-------- ( -21.80) >DroSim_CAF1 49233 102 + 1 UGUGUUUUUUUU--AACCGUGUAUUUUC--GC----UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU-------- ...........(--(((.(((......)--))----.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))))))))).)))).-------- ( -21.80) >DroEre_CAF1 60543 102 + 1 UGUGUUUUUUUU--AACCGUGUAUUUUC--GC----UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU-------- ...........(--(((.(((......)--))----.(((.((((((.(((.......((((((((((((.......))))))))))))..))).))))))))).)))).-------- ( -21.80) >DroYak_CAF1 76915 110 + 1 UGUGUUUUUUUU--AAAUGUGUAUUUUC--GC----UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUAUGUUGCUGUUUGUGCUA ............--....(((......)--))----.((((.((.(((((((((((..((((((((((((.......))))))))))))......)))).))))))).))..)))).. ( -24.70) >DroPer_CAF1 110547 94 + 1 ------UUUUUCGCUUUGGUGUACGGGCGGGCGGCAUACGCUGCCGCUGUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUAGCCAU------------------ ------..........((((.(((((((((((((((.....))))))).)))))))).((((((((((((.......))))))))))))......)))).------------------ ( -31.90) >consensus UGUGUUUUUUUU__AACCGUGUAUUUUC__GC____UGCAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU________ ..................(((((.............)))))((((((.(((.......((((((((((((.......))))))))))))..))).))))))................. (-15.96 = -16.07 + 0.11)

| Location | 7,300,345 – 7,300,447 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -12.82 |
| Consensus MFE | -8.31 |
| Energy contribution | -7.62 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7300345 102 - 22224390 --------ACAACAACACACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA----GC--GAAAAUACACGGUU--AAAAAAAACACA --------.............(((.....((((((((.(((.......))).)))))))).......(.(((((....))))----).--).........))).--............ ( -11.20) >DroSec_CAF1 50255 102 - 1 --------ACAACAACACACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA----GC--GAAAAUACACGGUU--AAAAAAAACACA --------.............(((.....((((((((.(((.......))).)))))))).......(.(((((....))))----).--).........))).--............ ( -11.20) >DroSim_CAF1 49233 102 - 1 --------ACAACAACACACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA----GC--GAAAAUACACGGUU--AAAAAAAACACA --------.............(((.....((((((((.(((.......))).)))))))).......(.(((((....))))----).--).........))).--............ ( -11.20) >DroEre_CAF1 60543 102 - 1 --------ACAACAACACACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA----GC--GAAAAUACACGGUU--AAAAAAAACACA --------.............(((.....((((((((.(((.......))).)))))))).......(.(((((....))))----).--).........))).--............ ( -11.20) >DroYak_CAF1 76915 110 - 1 UAGCACAAACAGCAACAUACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA----GC--GAAAAUACACAUUU--AAAAAAAACACA ..((.......(((......)))......((((((((.(((.......))).))))))))..........((((....))))----))--..............--............ ( -11.40) >DroPer_CAF1 110547 94 - 1 ------------------AUGGCUAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACACAGCGGCAGCGUAUGCCGCCCGCCCGUACACCAAAGCGAAAAA------ ------------------..(((......((((((((.(((.......))).))))))))...........((((((.....))))))..)))(((........))).....------ ( -20.70) >consensus ________ACAACAACACACUGCCAUAACAAUUAAUUAACAAAUAAAAUGUCAAUUAAUUUUACAAACAUUGCAGCGAUGCA____GC__GAAAAUACACGGUU__AAAAAAAACACA ..................((((.......((((((((.(((.......))).))))))))...........(((....)))..................))))............... ( -8.31 = -7.62 + -0.69)
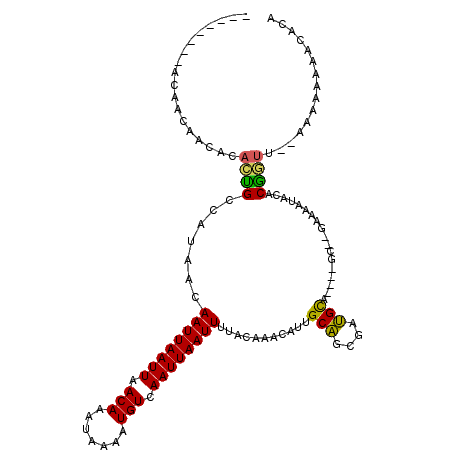
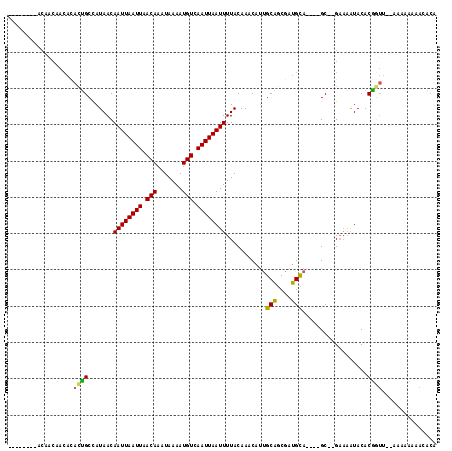
| Location | 7,300,375 – 7,300,471 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.07 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7300375 96 + 22224390 CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU---------UGUUCUUGUUAUUGUUUGGUUGAC ((.((((((.(((.......((((((((((((.......))))))))))))..))).)))))))).......---------........................ ( -18.00) >DroSec_CAF1 50285 90 + 1 CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU---------U------GUUAUUGUUUGGUUGGC ((.((((((.(((.......((((((((((((.......))))))))))))..))).)))))))).......---------.------................. ( -18.00) >DroSim_CAF1 49263 90 + 1 CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU---------U------GUUAUUGUUUGGUUGGC ((.((((((.(((.......((((((((((((.......))))))))))))..))).)))))))).......---------.------................. ( -18.00) >DroEre_CAF1 60573 96 + 1 CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU---------UGUUCUUGUUAUUGUUUGGUUGGC ((.((((((.(((.......((((((((((((.......))))))))))))..))).)))))))).......---------........................ ( -18.00) >DroYak_CAF1 76945 105 + 1 CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUAUGUUGCUGUUUGUGCUAUUGUUGUUGUUAUUGUUUGGUUGGC .((((..((((((.......((((((((((((.......))))))))))))((...((((((.....))))))...))............)))))).)))).... ( -20.40) >DroAna_CAF1 53558 82 + 1 ---CGCUGCAGUGUUUGUGAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCCGUGUGUUGUUGC--------------------CCGGUUGGUUGGC ---.(((.........((((((((((((((((.......)))))))))))).))))(((((.((......))--------------------.)))))....))) ( -18.10) >consensus CAUCGCUGCAAUGUUUGUAAAAUUAAUUGACAUUUUAUUUGUUAAUUAAUUGUUAUGGCAGUGUGUUGUUGU_________U______GUUAUUGUUUGGUUGGC ....((.((((..(((((..((((((((((((.......))))))))))))......)))).)..)))).))................................. (-16.45 = -16.07 + -0.39)

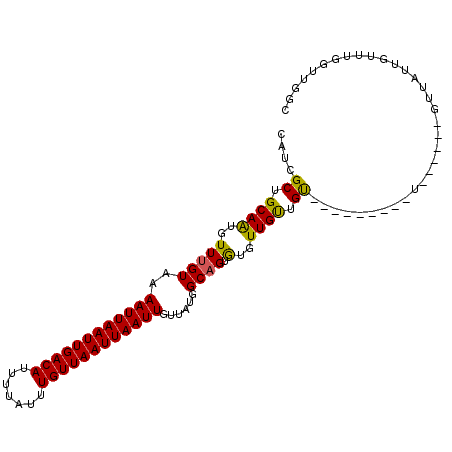
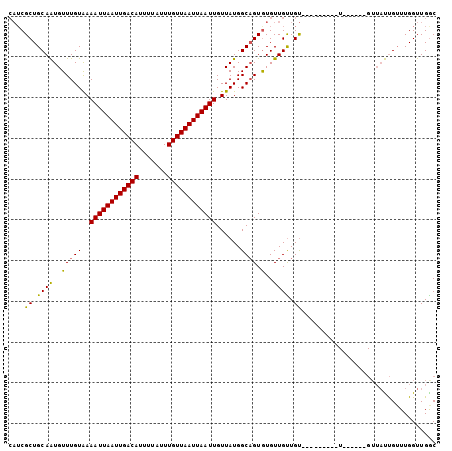
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:46 2006