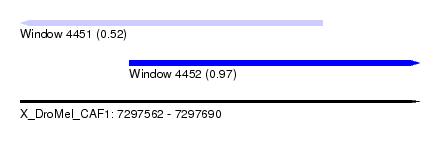
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,297,562 – 7,297,690 |
| Length | 128 |
| Max. P | 0.966983 |

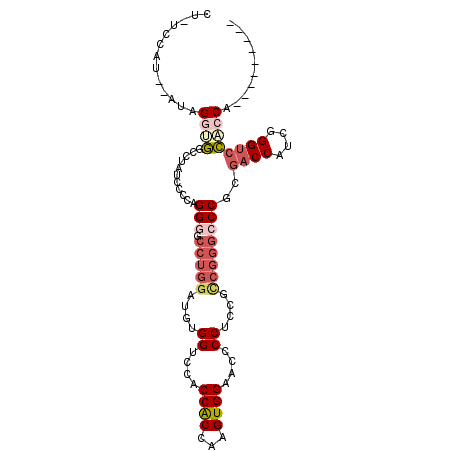
| Location | 7,297,562 – 7,297,659 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.46 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -13.24 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.46 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7297562 97 - 22224390 GGUGUUCCACUUGGUGGUGGACCACAUCCAGGCCCCUGGGGAUAGGCCACCUAU--GUAGAUGGCAGUUAUGUAUGUAUAAGUCACAAAUUAAUCAC-UA- .(((.((((((....)))))).))).(((((....)))))(((..((((.((..--..)).))))..(((((....)))))))).............-..- ( -26.50) >DroSec_CAF1 47410 87 - 1 GGGGUUCCACUUGGUGGUGGACCACAUCCAGGCCCCUGGGGAUAUGCCACCUAU--------AGCAGUUCC----AUAUAAGUCACAAAUUAAUCAC-UA- ..(((.(((((....))))))))....((((....))))(((..(((.......--------.)))..)))----......................-..- ( -23.00) >DroSim_CAF1 46376 93 - 1 GGGGUUCCACUUGGUGGUGGACCACAUCCAGGCCCCUGGGGAUAAGCCACCUAU--AAGGAUAGCAGUUAC----GCAUAAGUCACAAAUAAAUCAC-UA- ...(((...((((..(((((......(((((....)))))......)))))..)--)))...))).((.((----......)).))...........-..- ( -22.80) >DroEre_CAF1 57818 95 - 1 GGGGUUCCACUUGGCGGAGGACCGCAUCCAGGUCCCUGGGGAUGGGCCAGCUAA--GUGGAGAGCAGUUGA----UUAGAAGUCAGCAAUGCAUCCAAGGG .((((((((((((((...((.(((..(((((....)))))..))).)).)))))--)))))..((((((((----(.....))))))..)))))))..... ( -42.30) >DroYak_CAF1 74186 86 - 1 GGGGUGCCACUUGGUGGGGGGCCACAUCCAGGGCCCUGGGGAUAGGCCACCUAGUUAUGGACAGCAGUU--------AGA------GAAUGCAUCGCAUA- (.(((((..(((..((((.((((...(((((....)))))....)))).))))......(((....)))--------.))------)...))))).)...- ( -29.30) >DroPer_CAF1 107565 75 - 1 GGCGGGCCACUUGGUGGCGGUCCGCAGGGACCACCCUGGGGAUAAGCGCUCUUA--AAG--------------------AAAACAAAAAU--AUGAA-AA- ((((.(((((...))))).((((.(((((....))))).))))...))))....--...--------------------...........--.....-..- ( -27.10) >consensus GGGGUUCCACUUGGUGGUGGACCACAUCCAGGCCCCUGGGGAUAGGCCACCUAU__AUGGA_AGCAGUU_______UAUAAGUCACAAAUGAAUCAC_UA_ ((((((((.....((((....))))..((((....)))))))...))).)).................................................. (-13.24 = -13.88 + 0.64)

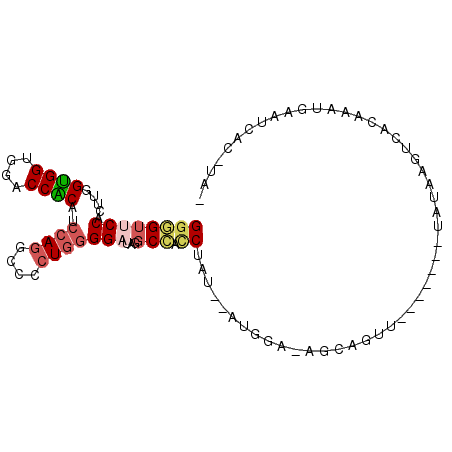
| Location | 7,297,597 – 7,297,690 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.83 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7297597 93 + 22224390 CCAUCUAC--AUAGGUGGCCUAUCCCCAGGGGCCUGGAUGUGGUCCACCACCAAGUGGAACACCUCCGUCGGGACCGCGACCAUCGGGUCCACCA--------- ........--...((((((((........))))).(((.(((.(((((......))))).))).)))....(((((.((.....)))))))))).--------- ( -37.60) >DroSec_CAF1 47441 87 + 1 CU--------AUAGGUGGCAUAUCCCCAGGGGCCUGGAUGUGGUCCACCACCAAGUGGAACCCCUGCGCCGGGCCCGCGACCAUCGGGUCCACCA--------- ..--------...((((............((((((((.((((((((((......)))).)))...)))))))))))..((((....)))))))).--------- ( -39.30) >DroSim_CAF1 46407 93 + 1 CUAUCCUU--AUAGGUGGCUUAUCCCCAGGGGCCUGGAUGUGGUCCACCACCAAGUGGAACCCCUCCGCCGGGCCCGCGACCAUCGGGUCCACCA--------- ........--...((((............((((((((..(.(((((((......)))).))).)....))))))))..((((....)))))))).--------- ( -38.70) >DroEre_CAF1 57851 93 + 1 CUCUCCAC--UUAGCUGGCCCAUCCCCAGGGACCUGGAUGCGGUCCUCCGCCAAGUGGAACCCCUCCGUCGGGCCCUCGACCAUCUGGUCCUCCA--------- ...(((((--((.((.(((((((((..((....))))))).)).))...)).)))))))..(((......))).....((((....)))).....--------- ( -32.90) >DroYak_CAF1 74208 95 + 1 CUGUCCAUAACUAGGUGGCCUAUCCCCAGGGCCCUGGAUGUGGCCCCCCACCAAGUGGCACCCCUCCCCCGGGCCCACGACCAUCUGGUCCUCCA--------- ....(((((.(((((.(.(((......))).)))))).)))))...........((((...(((......))).))))((((....)))).....--------- ( -32.20) >DroPer_CAF1 107583 97 + 1 -----CUU--UAAGAGCGCUUAUCCCCAGGGUGGUCCCUGCGGACCGCCACCAAGUGGCCCGCCGCCAUCUGGACCUCCGCCAUCGGGACGCCCGCCAGGCGGA -----...--((((....))))...((((((((((((....)))))))).....(((((.....)))))))))...((((((..((((...))))...)))))) ( -42.20) >consensus CU_UCCAU__AUAGGUGGCCUAUCCCCAGGGGCCUGGAUGUGGUCCACCACCAAGUGGAACCCCUCCGCCGGGCCCGCGACCAUCGGGUCCACCA_________ .............((((...........(((.(((((....((....((((...))))....))....))))))))..((((....)))))))).......... (-21.42 = -22.83 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:43 2006