
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,295,105 – 7,295,197 |
| Length | 92 |
| Max. P | 0.957992 |

| Location | 7,295,105 – 7,295,197 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7295105 92 - 22224390 -------------ACCACAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUACAAAAGGCAGAGUGCCAUGGCAAUGGCAACCAGGCCACCGAAA--------------- -------------.......((((......((((.....(((((........)))))..))))....(((....((((((....)))))).....))).))))..--------------- ( -20.90) >DroPse_CAF1 75300 106 - 1 -------------GCAGCAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUACAAAAGGCAGAGUGCCAUGGCAAUGGCAA-CAGGCCACCAAAUGGCACACCCGACUCU -------------..((.((((((...(((((((.....(((((........)))))..))))....(((....((((((....)))))).-...)))........)))...)))))))) ( -29.40) >DroEre_CAF1 55456 91 - 1 -------------ACCACAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUACAAAAGGCAGAGUGCCAUGGCAAUGGCAGCCAGGCCACCGAA---------------- -------------......(.((((.....((((.....(((((........)))))..))))....(((....((((((....)))))))))..)))).)...---------------- ( -22.10) >DroWil_CAF1 53270 110 - 1 CCCCCUUGAAGAAAAAACAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUAUAAAAGGCACAACGCCAUGGCAAUGGCAA-CAGGCCACGAAAUGGCAAA--------- ...................((((.((.(((....((.((((.......)))).))............(((.....)))...))).)).)))-)..((((.....))))...--------- ( -23.30) >DroAna_CAF1 48963 81 - 1 -------------ACCACAGUUGGCAAUGCGUAAAAGUGUUAACCGAAAGUAGUUAAAAUUACAAAAGGCAGAGCGCCAUGGCAAUGCCAG-----------AAA--------------- -------------.......((((((.(((((((.....((((((....)..)))))..))))....(((.....)))...))).))))))-----------...--------------- ( -21.90) >DroPer_CAF1 102991 106 - 1 -------------GUAGCAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUACAAAAGGCAGAGUGCCAUGGCAAUGGCAA-CAGGCCACCAAAUGGCACACCCGACUCU -------------..((.((((((...(((((((.....(((((........)))))..))))....(((....((((((....)))))).-...)))........)))...)))))))) ( -29.30) >consensus _____________ACAACAGUUGGCAAUGCGUAAAAGUGUUAACCAAAAGCAGUUAAAAUUACAAAAGGCAGAGUGCCAUGGCAAUGGCAA_CAGGCCACCAAAU_______________ ....................(((.((.(((((((.....(((((........)))))..))))....(((.....)))...))).)).)))............................. (-15.23 = -14.87 + -0.36)

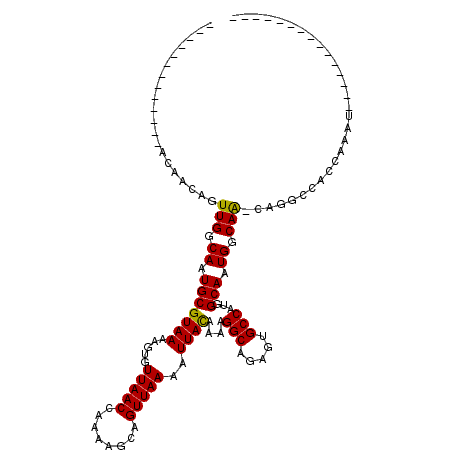
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:42 2006