
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,284,871 – 7,284,972 |
| Length | 101 |
| Max. P | 0.577815 |

| Location | 7,284,871 – 7,284,972 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -16.79 |
| Energy contribution | -17.98 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
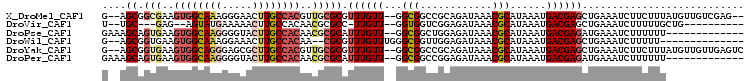
>X_DroMel_CAF1 7284871 101 - 22224390 G--AGCGGCGAAGUGGCAAAGGGAACUUGCCACGUUGCGCGUUUGUU--GGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUUUAUGUUGUCGAG-- .--....((((.(((((((.(....)))))))).))))(((((((((--.((....)).))))))))).....((((((..((((......))))..))))))..-- ( -36.70) >DroVir_CAF1 49286 87 - 1 U--UGC---GAG--AGUAUGAAAAACUUGCCACAACGCGCC-UUGUU--GGUGGUCGGAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUUUUGCUG---------- .--.((---(((--(..........((.(((((((((....-.))))--.))))).))((((....((...........))....))))))))))..---------- ( -15.50) >DroPse_CAF1 63865 92 - 1 GAAAGCAGUGAAGUGGCAAGGGGUACUUGCCACAACGCGCAUUUGUU--GGCGGCUGGAGAUAAACGCAUAAAUGACGAGAUGAAAUCUUUUUU------------- (((((..(((..((((((((.....))))))))..)))((((((((.--.(((..((....))..)))))))))).)..........)))))..------------- ( -26.70) >DroWil_CAF1 43000 89 - 1 G--AGCGGUGAAGUGGCAAAGGAAACUUGCCACAA--CGCGUUUGUUUGGGCGGUUGGAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUUUU-------------- (--(.((((...(((((((.(....))))))))..--.((((((((((..........))))))))))...........))))...)).....-------------- ( -28.60) >DroYak_CAF1 60936 103 - 1 G--AGCGGUGAAGUGGCAGGGAGCGCUUGCCACGUUGCGCGUUUGUU--GGCGGCCGCAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUCUUUAUGUUGUUGAGUC .--.(((((...((((((((.....))))))))....(((.......--.)))))))).(((.((((((((((.((.((.......)).)))))))).))))..))) ( -34.50) >DroPer_CAF1 91347 92 - 1 GAAAGCAGUGAAGUGGCAAGGGGUACUUGCCACAACGCGCAUUUGUU--GGCGGCCGGAGAUAAACGCAUAAAUGACGAGAUGAAAUCUUUUUU------------- (((((..(((..((((((((.....))))))))..)))((((((((.--.(((............)))))))))).)..........)))))..------------- ( -26.10) >consensus G__AGCGGUGAAGUGGCAAGGGGAACUUGCCACAACGCGCGUUUGUU__GGCGGCCGGAGAUAAACGCAUAAAUGACGAGCUGAAAUCUUUUUU_____________ ....((.(((..((((((((.....))))))))..))))).((((((...(((............)))......))))))........................... (-16.79 = -17.98 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:38 2006