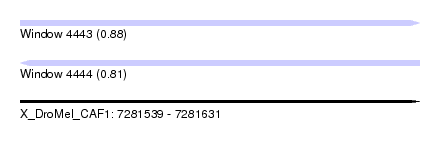
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,281,539 – 7,281,631 |
| Length | 92 |
| Max. P | 0.878424 |

| Location | 7,281,539 – 7,281,631 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 70.78 |
| Mean single sequence MFE | -16.65 |
| Consensus MFE | -11.39 |
| Energy contribution | -12.03 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
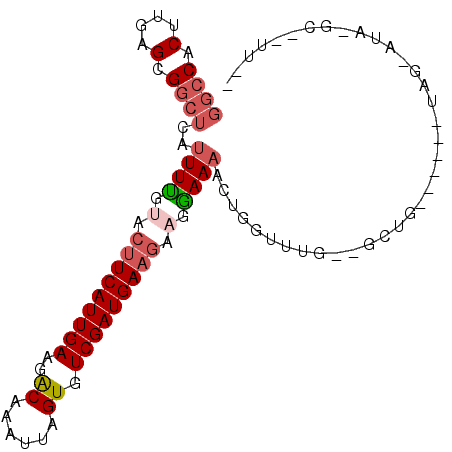
>X_DroMel_CAF1 7281539 92 + 22224390 CAAAGGGCUUAUACUA------CAAC--CAAACCAGUUUUCCUUCUUCAUCGACACUAAUUUGUCUUCAAUGAAGUACAAAAUGAGCCGCUCAAGUGGCC ....((..........------...)--)......(((((....((((((.((((......))))....))))))...)))))..(((((....))))). ( -16.52) >DroVir_CAF1 45833 89 + 1 -CAUGGAGAAUGGGCGCUCGCUCAGU--U----GAGCUUUUUGACUUCAUCGACACUAAUUUGUUUUCAAUGACUAGCAAA----CACACUCAAGUGGCG -....(((........)))((.((.(--(----(((...((((.((((((.((.((......))..)).))))..))))))----....))))).)))). ( -17.20) >DroSec_CAF1 31376 100 + 1 UAAAAAGCCUAUACUACUCGUACAACCCCUAACCAGUUUUCCUUCUUCAUCGACACUAAUUUGUCUUCAAUGAAGUACAAAAUGAGCCGCUCAAGUGGCC ...................................(((((....((((((.((((......))))....))))))...)))))..(((((....))))). ( -15.70) >DroSim_CAF1 31436 98 + 1 CGAAAAGCUUAUGCUACUCGUACAGC--CAAACCAAUUUUCCUUCUUCAUCGACACUAAUUUGUCUUCAAUGAAGUACGAAAUGAGCCGCUCAAGUGGCC (((..(((....)))..)))......--.........((((...((((((.((((......))))....))))))...))))...(((((....))))). ( -21.70) >DroEre_CAF1 41033 76 + 1 ----------------------CAGC--CAAACCAGUUUUCCUUCUUCAUCGACACUAAUUUGCCUUCAAUGAAGUACAAAAUGAGCCGCUCAAGUGGCC ----------------------....--.......(((((....((((((.((..(......)...)).))))))...)))))..(((((....))))). ( -13.10) >DroYak_CAF1 57500 76 + 1 ----------------------AAAA--AAAAACAGUUUUCCAUCUUCAUCGACACUAAUUUGUCUUCAAUGAAGUACAAAAUGAGCCGCUCAAGUGGCC ----------------------....--.......(((((....((((((.((((......))))....))))))...)))))..(((((....))))). ( -15.70) >consensus __AA__GC_UAU_CUA______CAAC__CAAACCAGUUUUCCUUCUUCAUCGACACUAAUUUGUCUUCAAUGAAGUACAAAAUGAGCCGCUCAAGUGGCC ...................................(((((....((((((.((((......))))....))))))...)))))..(((((....))))). (-11.39 = -12.03 + 0.64)



| Location | 7,281,539 – 7,281,631 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 70.78 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -13.55 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7281539 92 - 22224390 GGCCACUUGAGCGGCUCAUUUUGUACUUCAUUGAAGACAAAUUAGUGUCGAUGAAGAAGGAAAACUGGUUUG--GUUG------UAGUAUAAGCCCUUUG ((((.(....).))))..((((.(.(((((((((..((......)).))))))))).).))))...((((((--....------.....))))))..... ( -21.20) >DroVir_CAF1 45833 89 - 1 CGCCACUUGAGUGUG----UUUGCUAGUCAUUGAAAACAAAUUAGUGUCGAUGAAGUCAAAAAGCUC----A--ACUGAGCGAGCGCCCAUUCUCCAUG- (((((.((((((...----((((((..(((((((..((......)).))))))))).))))..))))----)--).)).))).................- ( -22.90) >DroSec_CAF1 31376 100 - 1 GGCCACUUGAGCGGCUCAUUUUGUACUUCAUUGAAGACAAAUUAGUGUCGAUGAAGAAGGAAAACUGGUUAGGGGUUGUACGAGUAGUAUAGGCUUUUUA (((((((((.(((((((.((((.(.(((((((((..((......)).))))))))).).)))).((....))))))))).)))))......))))..... ( -28.20) >DroSim_CAF1 31436 98 - 1 GGCCACUUGAGCGGCUCAUUUCGUACUUCAUUGAAGACAAAUUAGUGUCGAUGAAGAAGGAAAAUUGGUUUG--GCUGUACGAGUAGCAUAAGCUUUUCG .((.(((((.((((((((((((.(.(((((((((..((......)).))))))))).).))))..))....)--))))).))))).))............ ( -29.40) >DroEre_CAF1 41033 76 - 1 GGCCACUUGAGCGGCUCAUUUUGUACUUCAUUGAAGGCAAAUUAGUGUCGAUGAAGAAGGAAAACUGGUUUG--GCUG---------------------- (((((...((.(((....((((.(.(((((((((..((......)).))))))))).).)))).))).))))--))).---------------------- ( -21.10) >DroYak_CAF1 57500 76 - 1 GGCCACUUGAGCGGCUCAUUUUGUACUUCAUUGAAGACAAAUUAGUGUCGAUGAAGAUGGAAAACUGUUUUU--UUUU---------------------- ((((.(....).))))..(((..(.(((((((((..((......)).))))))))))..)))..........--....---------------------- ( -18.50) >consensus GGCCACUUGAGCGGCUCAUUUUGUACUUCAUUGAAGACAAAUUAGUGUCGAUGAAGAAGGAAAACUGGUUUG__GCUG______UAG_AUA_GC__UU__ ((((.(....).))))..((((.(.(((((((((..((......)).))))))))).).))))..................................... (-13.55 = -14.47 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:37 2006