
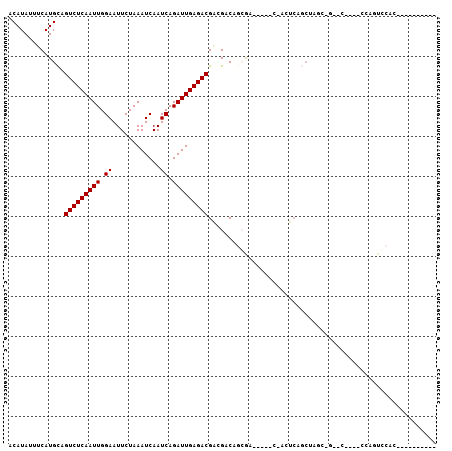
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,273,276 – 7,273,408 |
| Length | 132 |
| Max. P | 0.970519 |

| Location | 7,273,276 – 7,273,375 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -11.30 |
| Energy contribution | -11.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
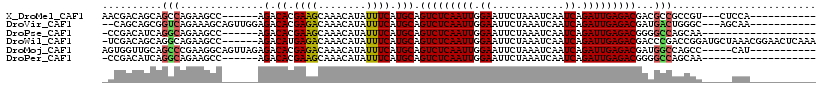
>X_DroMel_CAF1 7273276 99 - 22224390 ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACGCCGCCGU---CUCCACUCAGCUAACAGCUA---ACCAGUUCAGUUCUGUG--C ...........((((.((.((((((................((..((((((.......)))---)))...))(((.....))).---.)))))).))..)))).--. ( -22.10) >DroVir_CAF1 34737 85 - 1 ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGAUGACUGGGC---AGCAAC-----UGUCUGGCC---CACAGUCCA----------- ..............(((((((((.((............)).)))))))))(((...(((((---.((...-----.))...)))---))..)))..----------- ( -22.10) >DroPse_CAF1 50681 79 - 1 ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGGGGCCAGCAA--------CUCAGCUUG----------CCAAUGCAC---------- ..........(((((((((((((.((............)).)))))))))..(((.(((..--------....))).)----------))..)))).---------- ( -24.10) >DroGri_CAF1 30775 99 - 1 ACAUAUUUCAUGUAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGAUGACUGGGU---AGCAAUUCACCCGUCCGUCC---CACUGUCCACUUGUCUG--C ...........((((...(((.((((......((((.....))))(.((((..(((.((((---........))))))))))).---)....)))).))).)))--) ( -22.40) >DroAna_CAF1 25127 103 - 1 ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACGACGCCGGCUCCUCCACUCGGCCAGCAGUUCUCGGCCAGGUC----CUGAGUUU ..........((..(((((((((.((............)).)))))))))..)).....(((((....((..((((((.....)).))))..)).----..))))). ( -24.40) >DroPer_CAF1 77637 79 - 1 ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGGGGCCAGCAA--------CUCAGCUUG----------CCAAUGCAC---------- ..........(((((((((((((.((............)).)))))))))..(((.(((..--------....))).)----------))..)))).---------- ( -24.10) >consensus ACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACGACAGCGA_____C_ACUCAGCUAGC_G__C____CCAGUCCAC__________ ..............(((((((((.((............)).)))))))))......................................................... (-11.30 = -11.30 + -0.00)


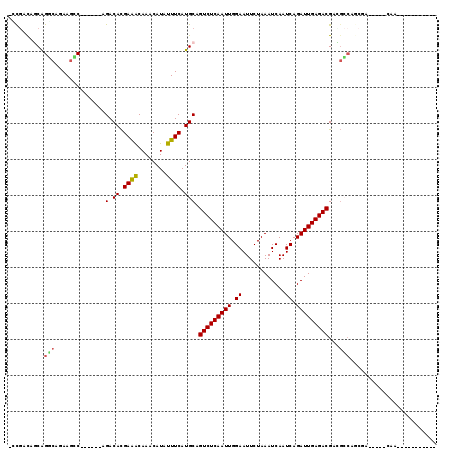
| Location | 7,273,309 – 7,273,408 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
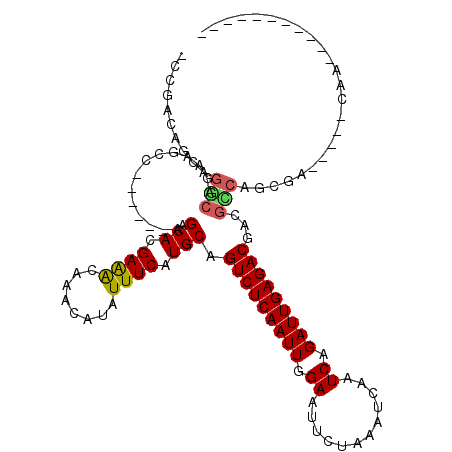
>X_DroMel_CAF1 7273309 99 - 22224390 AACGACAGCAGCCAGAAGCC------AGACACGAAGCAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACGCCGCCGU---CUCCA----------- ...(((.((.((........------.........(((...........))).(((((((((.((............)).)))))))))...)).)).))---)....----------- ( -21.00) >DroVir_CAF1 34756 103 - 1 --CAGCAGCGGUCAGAAAGCAGUUGGAGACACGAGACAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGAUGACUGGGC---AGCAA----------- --..((..((((((....(((((.(....)))((((........)))).))).(((((((((.((............)).)))))))))..)))))).))---.....----------- ( -28.40) >DroPse_CAF1 50699 93 - 1 -CCGACAUCAGGCAGAAGCC------AGACACGAAGCAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGGGGCCAGCAA------------------- -.........(((....)))------......((((........)))).(((.(((((((((.((............)).)))))))))(....).))).------------------- ( -18.90) >DroWil_CAF1 30738 112 - 1 -UCGACAGCAGGCAGAAGCC------AGACAUGAGACAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACCCGACCGGAUGCUAAACGGAACUCAAA -(((..(((((((....)))------.(.(((((((........)))))))).(((((((((.((............)).)))))))))...((....)).))))...)))........ ( -26.50) >DroMoj_CAF1 38322 103 - 1 AGUGGUUGCAGCCCGAAGGCAGUUAGAGACACGAGACAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGAUGGCCAGCC-----CAU----------- .(.(((((..(((.....((((((...)))..((((........)))).))).(((((((((.((............)).)))))))))...))))))))-----)..----------- ( -24.50) >DroPer_CAF1 77655 93 - 1 -CCGACAUCAGGCAGAAGCC------AGACACGAAGCAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGGGGCCAGCAA------------------- -.........(((....)))------......((((........)))).(((.(((((((((.((............)).)))))))))(....).))).------------------- ( -18.90) >consensus _CCGACAGCAGGCAGAAGCC______AGACACGAAACAAACAUAUUUCAUGCAGUCUCAAUUGGAAUUCUAAAUCAAUCAGAUUGAGACGACGCCAGCGA_____CAA___________ ..........(((..............(.((.((((........)))).))).(((((((((.((............)).)))))))))...)))........................ (-14.77 = -14.38 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:30 2006