
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,264,001 – 7,264,207 |
| Length | 206 |
| Max. P | 0.768932 |

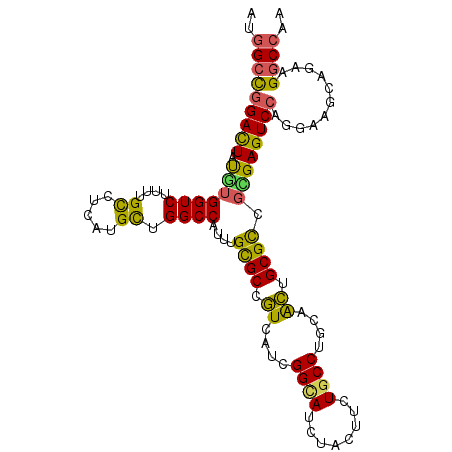
| Location | 7,264,001 – 7,264,106 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -25.49 |
| Energy contribution | -24.13 |
| Covariance contribution | -1.35 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7264001 105 + 22224390 AUGGCCGGAUUAUUUGGUCUUUUGCCUGAUGUUGGCCAUUUGCGCCGUUAUCGGUAUAUAUUUUUGCCUGCAACUGCGCUUAGAGUCCAGGAAACAAAAGGCCAA .((((((((((....((((..((((..((((.(((((....).)))).))))((((........)))).))))..).)))...))))).(....)....))))). ( -35.20) >DroVir_CAF1 22327 105 + 1 UUGGCCAGACUAUGUGGUCUUUUGUCUCAUGCUGGCCAUCUGCGCCGUCAUCGGCAUCUAUUUCUGCCUGCAGCUGCGUCGCGAGUCGAAGGAGGUGAAGGCCUC ..(((((((((....)))))((..((((.(((.(((.......((((....))))..........))).))).((.((.(....).)).))))))..)))))).. ( -36.63) >DroGri_CAF1 18935 105 + 1 CUGGCCGGACUAUCUGGUCUUUAGCCUCAUGCUGGCCAUUUGCGCCUUUAUCGGCAUCUACUUUUGCCUGCAGAUGCGCCGGGAGUCCAGACAGCUCAAGGCCAG ((((((.((((.(((((.(((((((.....)))(((((((((((........((((........)))))))))))).))).)))).))))).)).))..)))))) ( -44.30) >DroEre_CAF1 23690 105 + 1 AUGGCUGGACUACUUGGUCUUCUGCCUGAUGCUGGCCAUUUGCGCCGUGAUCGGGAUUUAUUUCUGCCUGCAACUGCGUGGCGAGUCCAGGAGGAAGAAGGCGCA (((((..(.(.....(((.....)))....))..))))).((((((....((.((((((......((((((....))).)))))))))..)).......)))))) ( -35.40) >DroYak_CAF1 38806 105 + 1 AUGGCCGGAUUAUGUGGUCUUUUGCCUGAUGUUGGCCAUUUGUGCUGUAAUUGGCAUUUACUUUUGCCUGCAACUGCGUGGCGAGUCCAGGAAAAAGAAGUCCUU ((((((((((((.(..(....)..).)))).))))))))..(((((......))))).....(((((((((....))).))))))...((((........)))). ( -30.00) >DroMoj_CAF1 22047 105 + 1 CUGGCCGGACUACGUGGUCUUCUGCCUCAUGCUGGCCAUCUGCGCCGUCAUCGGCAUCCACUUCUGCCUGCAGCUGCGCCGCGAGUCCCGCCAGCUGAAGGCCAC (((((.(((((.(((((((..((((...(((.(((((....).)))).))).((((........)))).))))..).))))))))))).)))))........... ( -43.60) >consensus AUGGCCGGACUAUGUGGUCUUUUGCCUCAUGCUGGCCAUUUGCGCCGUCAUCGGCAUCUACUUCUGCCUGCAACUGCGCCGCGAGUCCAGGAAGCAGAAGGCCAA ..(((((((((.(((((((....((.....)).))))....((((.((....((((........))))....)).)))).))))))))...........)))).. (-25.49 = -24.13 + -1.35)

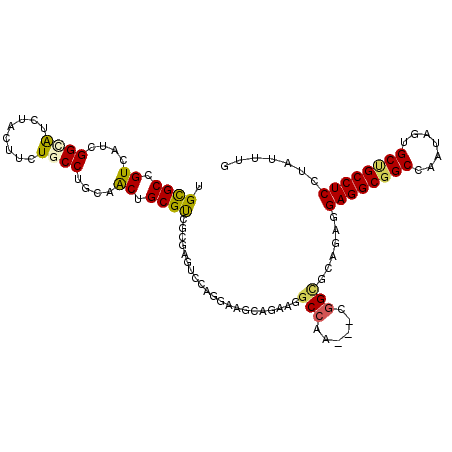
| Location | 7,264,041 – 7,264,146 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -26.41 |
| Energy contribution | -26.13 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7264041 105 + 22224390 UGCGCCGUUAUCGGUAUAUAUUUUUGCCUGCAACUGCGCUUAGAGUCCAGGAAACAAAAGGCCAA---UGGCUCAGAGGAGGCGGCGAGUAGUGCUGCCUCGUAUUUG .((((.(((...((((........))))...))).))))...(((((..((...(....).))..---.)))))....(((((((((.....)))))))))....... ( -35.40) >DroVir_CAF1 22367 105 + 1 UGCGCCGUCAUCGGCAUCUAUUUCUGCCUGCAGCUGCGUCGCGAGUCGAAGGAGGUGAAGGCCUC---CGGUGGCGAGGAGGCGGCCAAUAGCGCCGCCUCCUAUUUG .....(((((((((((........))))........((.(....).))..((((((....)))))---))))))))(((((((((((....).))))))))))..... ( -50.20) >DroEre_CAF1 23730 105 + 1 UGCGCCGUGAUCGGGAUUUAUUUCUGCCUGCAACUGCGUGGCGAGUCCAGGAGGAAGAAGGCGCA---UGGCGAAGAGGAGGCGGCGACUAGUGCUGCCUCAUAUUUG ((((((....((.((((((......((((((....))).)))))))))..)).......))))))---...((((...(((((((((.....)))))))))...)))) ( -41.30) >DroYak_CAF1 38846 105 + 1 UGUGCUGUAAUUGGCAUUUACUUUUGCCUGCAACUGCGUGGCGAGUCCAGGAAAAAGAAGUCCUU---CGGCGAAGAGGAGGCGGCCACUAGUGCUGCCUCGUAUUUG .(((((......)))))...(((((((((((....))).)))..(((.((((........)))).---.))).)))))(((((((((....).))))))))....... ( -35.40) >DroMoj_CAF1 22087 105 + 1 UGCGCCGUCAUCGGCAUCCACUUCUGCCUGCAGCUGCGCCGCGAGUCCCGCCAGCUGAAGGCCAC---CGGCGCCGAGGAGGCGGCCAGCAGCGCCGCCUCCUACCUG .((((((.....((...))......((((.((((((((.(....)...)).)))))).))))...---))))))..(((((((((((....).))))))))))..... ( -52.00) >DroPer_CAF1 67355 108 + 1 UGCGCCGUGAUUGGGGUCUACUUCUGCCUGCAGCUGCGUCGAGAGUCCCGCAAGCAGCGGGCCCACGGUGGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUG ..(((((((...((((....)))).((((((.((((((.(....)...))).))).)))))).)))))))......(((((((.(((....).)).)))))))..... ( -48.90) >consensus UGCGCCGUCAUCGGCAUCUACUUCUGCCUGCAACUGCGUCGCGAGUCCAGGAAGCAGAAGGCCAA___CGGCGCAGAGGAGGCGGCCAAUAGUGCUGCCUCCUAUUUG .((((.((....((((........))))....)).)))).....................(((......)))......((((((((.......))))))))....... (-26.41 = -26.13 + -0.27)


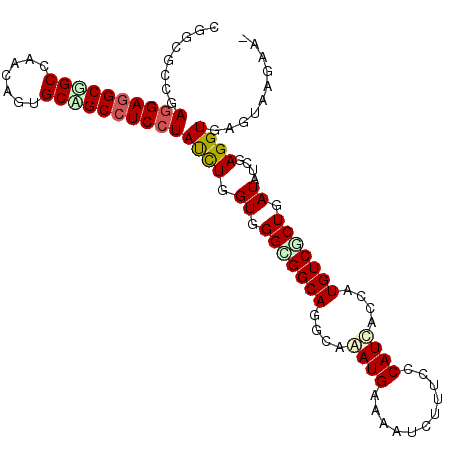
| Location | 7,264,106 – 7,264,207 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7264106 101 + 22224390 UGGCUCAGAGGAGGCGGCGAGUAGUGCUGCCUCGUAUUUGGUUGGUGGCAGGCAGAUGAAGAUCUUUCCCAUAACCAUGUCACUGAUAUCCAGGUGAGGAA---- ...(((....(((((((((.....)))))))))..(((((((..((((((((...(((..((....)).)))..)).))))))..)...)))))))))...---- ( -36.00) >DroVir_CAF1 22432 105 + 1 CGGUGGCGAGGAGGCGGCCAAUAGCGCCGCCUCCUAUUUGGUCGGCGGCAGAAAAAUGAAAAUCUUUCCCAUUACCAUGUCGCUAAUCUCUAGGUGAGUACGAAA .((((((((((((((((((....).))))))))))...((((.(..((..((((.((....)).)))))).).)))))))))))...(((.....)))....... ( -37.40) >DroPse_CAF1 40897 105 + 1 UGGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUAGUGGGCGGCAGGCAGAUGAAGAUAUUUCCCAUCACCAUGUCUCUGAUAUCGAGGUGGGUAUGAAU (((((((.....))).)))).((.(((.(((.(((((...))))).)))..)))..)).........((((((.(.(((((...))))).).))))))....... ( -35.60) >DroWil_CAF1 21575 101 + 1 CAGUGCAGAGGAGGCAGCUGAUAGUGCUGCAUCCUAUCUGGUGGGUGGCAGGCAAAUGAAAAUCUUUCCCAUUACCAUGUCGCUGAUAGCAAGGUGAGUUA---- (..(((..((((.(((((.......))))).)))).....((.(((((((((..((((...........)))).)).))))))).)).)))..).......---- ( -30.20) >DroMoj_CAF1 22152 105 + 1 CGGCGCCGAGGAGGCGGCCAGCAGCGCCGCCUCCUACCUGGUGGGCGGCAGGAAAAUGAAAAUCUUUCCCAUCACCAUGUCGCUUAUCUCGAGGUGAGUACAAAA ...((((.(((((((((((....).))))))))))....((((((((((((((((((....)).)))))........)))))))))))....))))......... ( -44.90) >DroPer_CAF1 67423 105 + 1 UGGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUGGUGGGCGGCAGGCAGAUGAAGAUAUUUCCCAUCACCAUGUCUCUGAUAUCGAGGUGGGUAUGAAU (((((((.....))).)))).((.(((.(((.(((((...))))).)))..)))..)).........((((((.(.(((((...))))).).))))))....... ( -35.60) >consensus CGGCGCCGAGGAGGCGGCCAACAGUGCAGCCUCCUAUCUGGUGGGCGGCAGGCAAAUGAAAAUCUUUCCCAUCACCAUGUCGCUGAUAUCGAGGUGAGUAAGAA_ ........((((((((((.......))))))))))((((.((.(((((((....((((...........))))....))))))).))....)))).......... (-27.03 = -27.40 + 0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:27 2006