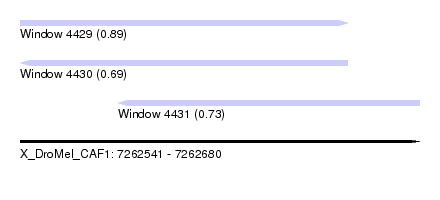
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,262,541 – 7,262,680 |
| Length | 139 |
| Max. P | 0.891976 |

| Location | 7,262,541 – 7,262,655 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -28.41 |
| Energy contribution | -30.67 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
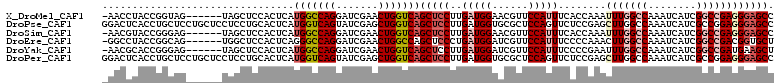
>X_DroMel_CAF1 7262541 114 + 22224390 CUUGUGUCCUAUUUAUGGGUCAGAGCUGCUUGGCGGCUCCCUCGGCCGAUGAUUUGGCCAAAUUUGGUGAAAUGGAACGUUCCAUCAAGGAGCUGACCAGUUCGAUCCUGGCCA ................(((((.((((((.....(((((((...((((((....))))))....((((((.((((...)))).)))))))))))))..)))))))))))...... ( -42.20) >DroPse_CAF1 39509 114 + 1 CUCUAUUUUUGUGCUUGACCCAGAACUGCUGGGCGGCUCCCUCCGGCGAUGAUUUGGCCAAGCUCGGAGAACUGGAGCGCACCAUCAAGGAGCUGACCAGCUCGAUACUGACCA ..........((((.((.(((((.....)))))))((((((((((((..((.......)).)).)))))....))))))))).......(((((....)))))........... ( -39.00) >DroSim_CAF1 11223 114 + 1 CUUGUAUCCUAUUUAUGGGCCAGAAUUGCUUGGCGGCUCCCUCGGCCGAUGAUUUGGCCAAAUUUGGUGAAAUGGAACGUUCCAUCAAGGAGCUGACCAGUUCGAUCCUGGCCA .................((((((.((((.(((((((((((...((((((....))))))....((((((.((((...)))).))))))))))))).))))..)))).)))))). ( -47.90) >DroYak_CAF1 37212 114 + 1 UUUGUAUCCUGUUUAUGGGUCAGCAUUGUUGGGCAGCUUCAUCGGCCGAUGAUUUGGCCAAAUUCGGGGAAAUGGAACGAUCCAUCAAGGAGCUGACCAGUUCGAUCCUGGCCA .................((((((.((((((((.(((((((...((((((....))))))............(((((....)))))...))))))).))))..)))).)))))). ( -41.70) >DroAna_CAF1 14769 114 + 1 UAUUGGCCCUGUGCCUGGGCCACAGUGUGUGGGCAGCUCCCUCGGCGGAGGACCUGGCCCAGUUCGGGGAGCUGGAGAAAACCAUCAAGGAGCUAACCAACUCGAUCUUGGCCA ....((.(((.((((.(((((((.....))))......)))..)))).))).)).((((.((.(((((..(.(((......))).)..((......))..))))).)).)))). ( -42.40) >DroPer_CAF1 66055 114 + 1 CUCUAUUUUUGUGCUUGACCCAGAACUGCUGGGCGGCUCCCUCCGGCGAUGAUUUGGCCAAGCUCGGAGAACUGGAGCGCACCAUCAAGGAGCUGACCAGCUCGAUACUGACCA ..........((((.((.(((((.....)))))))((((((((((((..((.......)).)).)))))....))))))))).......(((((....)))))........... ( -39.00) >consensus CUUGUAUCCUGUGCAUGGGCCAGAACUGCUGGGCGGCUCCCUCGGCCGAUGAUUUGGCCAAAUUCGGAGAAAUGGAACGAACCAUCAAGGAGCUGACCAGCUCGAUCCUGGCCA .................((((((....(((((.(((((((...((((((....)))))).........((..(((......)))))..))))))).)))))......)))))). (-28.41 = -30.67 + 2.25)


| Location | 7,262,541 – 7,262,655 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.55 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7262541 114 - 22224390 UGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGUUCCAUUUCACCAAAUUUGGCCAAAUCAUCGGCCGAGGGAGCCGCCAAGCAGCUCUGACCCAUAAAUAGGACACAAG (((((((.......)))))))(.((((..((((...................(((((((........)))))))(((((.((...)).)))))...))))....)))))..... ( -36.90) >DroPse_CAF1 39509 114 - 1 UGGUCAGUAUCGAGCUGGUCAGCUCCUUGAUGGUGCGCUCCAGUUCUCCGAGCUUGGCCAAAUCAUCGCCGGAGGGAGCCGCCCAGCAGUUCUGGGUCAAGCACAAAAAUAGAG ((..((((.....))))..)).(((..(....(((((((((....(((((.((.(((.....)))..)))))))))))).((((((.....))))))...))))....)..))) ( -41.70) >DroSim_CAF1 11223 114 - 1 UGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGUUCCAUUUCACCAAAUUUGGCCAAAUCAUCGGCCGAGGGAGCCGCCAAGCAAUUCUGGCCCAUAAAUAGGAUACAAG .(((((((((.(...((((..(((((((((((((....))))))..........(((((........)))))))))))).))))..).)))))))))................. ( -43.20) >DroYak_CAF1 37212 114 - 1 UGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAUCGUUCCAUUUCCCCGAAUUUGGCCAAAUCAUCGGCCGAUGAAGCUGCCCAACAAUGCUGACCCAUAAACAGGAUACAAA (((((((.......)))))))..((((..((((.((.................((((((........))))))...(((...........))))).))))....))))...... ( -30.30) >DroAna_CAF1 14769 114 - 1 UGGCCAAGAUCGAGUUGGUUAGCUCCUUGAUGGUUUUCUCCAGCUCCCCGAACUGGGCCAGGUCCUCCGCCGAGGGAGCUGCCCACACACUGUGGCCCAGGCACAGGGCCAAUA (((((.((.(((((((((..((..((.....))..))..)))))))...)).)).)))))((((((..(((..(((......((((.....))))))).)))..)))))).... ( -47.10) >DroPer_CAF1 66055 114 - 1 UGGUCAGUAUCGAGCUGGUCAGCUCCUUGAUGGUGCGCUCCAGUUCUCCGAGCUUGGCCAAAUCAUCGCCGGAGGGAGCCGCCCAGCAGUUCUGGGUCAAGCACAAAAAUAGAG ((..((((.....))))..)).(((..(....(((((((((....(((((.((.(((.....)))..)))))))))))).((((((.....))))))...))))....)..))) ( -41.70) >consensus UGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGCUCCAGUUCACCGAACUUGGCCAAAUCAUCGGCCGAGGGAGCCGCCCAGCAAUUCUGGCCCAUAAACAGGAAAAAAG (((((((.......)))))))(((((....(((......)))..........(((((((........))))))))))))................................... (-23.02 = -23.55 + 0.53)
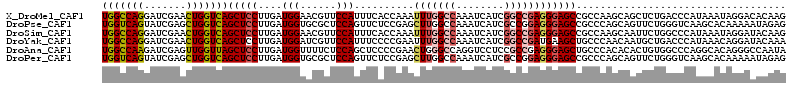
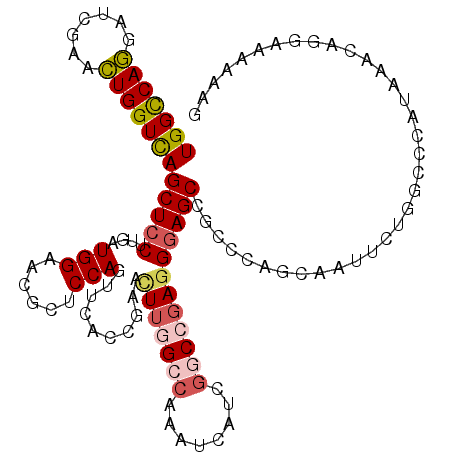

| Location | 7,262,575 – 7,262,680 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.65 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -24.07 |
| Energy contribution | -25.43 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7262575 105 - 22224390 -AACCUACCGGUAG------UAGCUCCACUCAUGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGUUCCAUUUCACCAAAUUUGGCCAAAUCAUCGGCCGAGGGAGCC -.(((....)))..------..(((((.(((.(((((((.......))))))).......((((((....))))))...........((((........)))))))))))). ( -38.00) >DroPse_CAF1 39543 112 - 1 GGACUCACCUGCUCCUGCUCCUCCUGCACUCAUGGUCAGUAUCGAGCUGGUCAGCUCCUUGAUGGUGCGCUCCAGUUCUCCGAGCUUGGCCAAAUCAUCGCCGGAGGGAGCC ((.(((....((....)).(((((.((.....(((((((..(((((((((...((.((.....)).))...)))))))...))..))))))).......)).)))))))))) ( -41.90) >DroSim_CAF1 11257 105 - 1 -AACGUACCGGGAG------UAGCUCCACUCAUGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGUUCCAUUUCACCAAAUUUGGCCAAAUCAUCGGCCGAGGGAGCC -.........((((------...)))).....(((((((.......)))))))(((((((((((((....))))))..........(((((........)))))))))))). ( -37.90) >DroEre_CAF1 22387 105 - 1 -GGCCUACCGGCAG------UGGCUCCACUCAGGGCCAGGAUCGAACUGGCCAGCUCCCUGAUGGAUCGUUCCAUUUCCCCAAACUUGGCCAAAUCAUCGGCCGACGGUGCU -.(((....)))((------((....))))...((((((.......))))))(((.((..((((((....)))))).........((((((........)))))).)).))) ( -39.80) >DroYak_CAF1 37246 105 - 1 -AACGCACCGGGAG------UAGCUCCACUCAUGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAUCGUUCCAUUUCCCCGAAUUUGGCCAAAUCAUCGGCCGAUGAAGCU -...((..((((((------.((((........((((((.......)))))))))).)).((((((....))))))..))))...((((((........))))))....)). ( -35.20) >DroPer_CAF1 66089 112 - 1 GGACUCACCUGCUCCUGCUCCUCCUGCACUCAUGGUCAGUAUCGAGCUGGUCAGCUCCUUGAUGGUGCGCUCCAGUUCUCCGAGCUUGGCCAAAUCAUCGCCGGAGGGAGCC ((.(((....((....)).(((((.((.....(((((((..(((((((((...((.((.....)).))...)))))))...))..))))))).......)).)))))))))) ( -41.90) >consensus _AACCCACCGGCAG______UAGCUCCACUCAUGGCCAGGAUCGAACUGGUCAGCUCCUUGAUGGAACGUUCCAUUUCACCAAACUUGGCCAAAUCAUCGGCCGAGGGAGCC ................................(((((((.......)))))))(((((..(((((......)))))........(((((((........)))))))))))). (-24.07 = -25.43 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:24 2006