
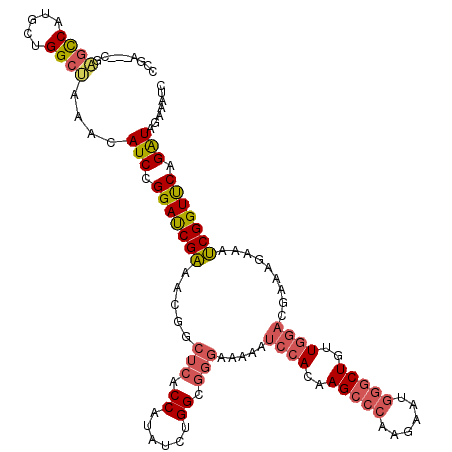
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,252,858 – 7,253,015 |
| Length | 157 |
| Max. P | 0.874071 |

| Location | 7,252,858 – 7,252,975 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -25.69 |
| Energy contribution | -27.00 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7252858 117 + 22224390 CCGA---CGAGCCAUGCUGGCUAAACAUCCGGAUCGAAACGGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGCUGGAGGGAAGAAAUCGGUUCAGAUAGAAAUC ((((---.(((((...((((........))))........))))).((...((..((.(((....)))...((((((....))))))))..)).))......)))).............. ( -37.60) >DroSec_CAF1 3103 117 + 1 CCGA---CGAGUCAUGCUGGCUAAAGAUCUGGAUCGAAGCGGCUCACCAUAUCUGGCAGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACGAAAGAAAUCGGUUCAGAUAGAAAUC ....---..((((.....))))....(((((((((((...((....))...((..((.(((....)))...((((((....))))))))..))(....)...)))))))))))....... ( -36.80) >DroSim_CAF1 1545 117 + 1 CCGA---CGUUCCAUGCUGGCUAAAGAUCCGGAUCGAAACGGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACUAAAGAAAUCGGUUCAGAUAGAAAUC ((((---..((((.(((..(.((..((.(((........))).))....)).)..)))))))...((((..((((((....))))))..)))).........)))).............. ( -33.70) >DroAna_CAF1 5088 118 + 1 CCACAGGCAGGUCAUGCUGGCCAACCAUCCGGACCGCUGUGGCUCUCCCUAUUUGGCGUCCAAAAUCAACGAGGCCAAGGAGCUGCUAAUGUCGCGGAGGCAGCGGUCCCGGUAGGGA-- ((...((..((((.....))))..))(((.((((((((((....((((...((((((.((..........)).))))))..((.((....)).)))))))))))))))).))).))..-- ( -48.20) >consensus CCGA___CGAGCCAUGCUGGCUAAACAUCCGGAUCGAAACGGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACGAAAGAAAUCGGUUCAGAUAGAAAUC .........((((.....))))....(((.(((((((.....(((.((......)).))).....((((..(((((......)))))..)))).........))))))).)))....... (-25.69 = -27.00 + 1.31)


| Location | 7,252,895 – 7,253,015 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.07 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -28.01 |
| Energy contribution | -28.01 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7252895 120 + 22224390 GGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGCUGGAGGGAAGAAAUCGGUUCAGAUAGAAAUCGAAAGAUUAGGCACUAGAUGGCAGGAUGUGAAGUUCAACA ......((...((..((.(((....)))...((((((....))))))))..)).))..(((.(((((((.......((((....))))..((.(.....))).)))).)))..))).... ( -31.90) >DroSec_CAF1 3140 113 + 1 GGCUCACCAUAUCUGGCAGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACGAAAGAAAUCGGUUCAGAUAGAAAUCGAAAGAUUAGG-------UGGCAGGAAUUGAAGUUCAACA .((.((((.((((((((..((....((((..((((((....))))))..))))(....)...)).).)))))))..((((....)))).))-------))))..((((....)))).... ( -38.60) >DroSim_CAF1 1582 113 + 1 GGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACUAAAGAAAUCGGUUCAGAUAGAAAUCGAAAGAUUAGG-------UGGCAGGAAUUGAAGUUCAACA .((.((((.((((((((((......((((..((((((....))))))..)))).........)))..)))))))..((((....)))).))-------))))..((((....)))).... ( -36.66) >consensus GGCUCACCAUAUCUGGCGGGAAAAAUCCACAAGCCCAAGAAUGGGCUGUUGGACGAAAGAAAUCGGUUCAGAUAGAAAUCGAAAGAUUAGG_______UGGCAGGAAUUGAAGUUCAACA ..((..(((((((((((.((.....((((..((((((....))))))..)))).........)).).)))))))..((((....))))..........)))..))............... (-28.01 = -28.01 + 0.00)



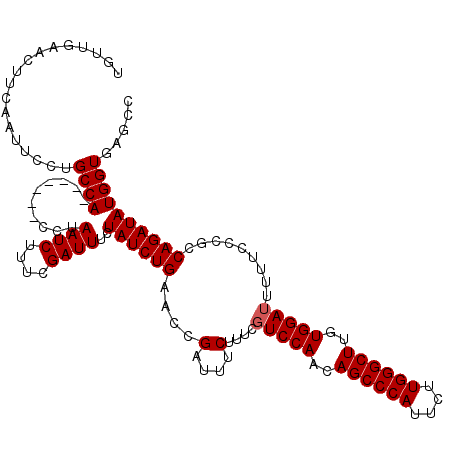
| Location | 7,252,895 – 7,253,015 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.07 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.680740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7252895 120 - 22224390 UGUUGAACUUCACAUCCUGCCAUCUAGUGCCUAAUCUUUCGAUUUCUAUCUGAACCGAUUUCUUCCCUCCAGCAGCCCAUUCUUGGGCUUGUGGAUUUUUCCCGCCAGAUAUGGUGAGCC .(..(((.((((((...(((............((((.((((((....))).)))..))))...........)))(((((....))))).)))))).)))..)(((((....))))).... ( -25.40) >DroSec_CAF1 3140 113 - 1 UGUUGAACUUCAAUUCCUGCCA-------CCUAAUCUUUCGAUUUCUAUCUGAACCGAUUUCUUUCGUCCAACAGCCCAUUCUUGGGCUUGUGGAUUUUUCCUGCCAGAUAUGGUGAGCC .((((.....))))....((((-------((.((((....))))..((((((...(((......)))((((..((((((....))))))..))))..........)))))).)))).)). ( -31.30) >DroSim_CAF1 1582 113 - 1 UGUUGAACUUCAAUUCCUGCCA-------CCUAAUCUUUCGAUUUCUAUCUGAACCGAUUUCUUUAGUCCAACAGCCCAUUCUUGGGCUUGUGGAUUUUUCCCGCCAGAUAUGGUGAGCC .((((.....))))....((((-------((.((((....))))..((((((...((........((((((..((((((....))))))..)))))).....)).)))))).)))).)). ( -30.22) >consensus UGUUGAACUUCAAUUCCUGCCA_______CCUAAUCUUUCGAUUUCUAUCUGAACCGAUUUCUUUCGUCCAACAGCCCAUUCUUGGGCUUGUGGAUUUUUCCCGCCAGAUAUGGUGAGCC ..................((((..........((((....))))..((((((....(....)....(((((..((((((....))))))..))))).........))))))))))..... (-23.00 = -23.33 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:21 2006