
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,224,760 – 7,224,895 |
| Length | 135 |
| Max. P | 0.641431 |

| Location | 7,224,760 – 7,224,857 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -14.17 |
| Energy contribution | -15.25 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
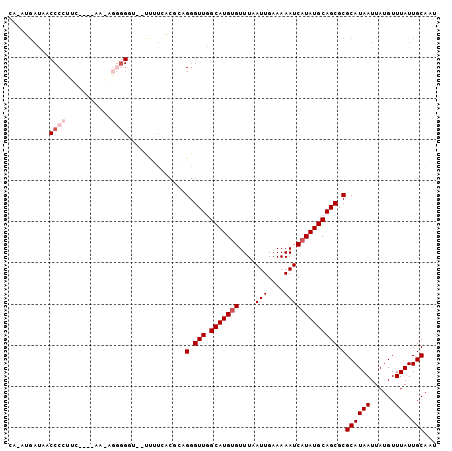
>X_DroMel_CAF1 7224760 97 + 22224390 CA-AUGAUAACCCCUCA----AAGAGGGGGU--UUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU ..-.(((.(((((((..----....))))))--)..))).((((.((((.((((((....(((....))))))))))))).)((((....))))....)))... ( -27.20) >DroPse_CAF1 1503 78 + 1 ------------CCUUCG-----------UG--CAGCAGCAGCGGGUU-GCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU ------------......-----------((--(((.(((((((.(((-(((((((....(((....))))))))))))).))).......))))..))))).. ( -23.81) >DroSim_CAF1 1266 97 + 1 CA-AUGAUAACCCCUCA----AAGAGGGGAU--UUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU ((-((((...((((((.----..))))))..--.......((.(.((((.((((((....(((....))))))))))))).)))..........)))))).... ( -26.20) >DroEre_CAF1 1236 97 + 1 CA-AUGAUAGCCCCUUC----AAAAGGGGGC--UUUUCACGCCGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU ..-.(((.(((((((..----....))))))--)..)))....(.((((.((((((....(((....))))))))))))).)((((((......))).)))... ( -28.90) >DroYak_CAF1 1944 104 + 1 CAAAUGAUAACCCCUUCGCAAAAACGGGGGCCUUUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU .......(((((((((((......)))))((.........)).))))))(((((((....(((....))))))))))...(((.((((......)))))))... ( -22.40) >DroAna_CAF1 1233 88 + 1 GGAACGAC--CCCCUUCG----------GCU--CCGUGGCAACGGGUU-GCAUG-GUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU (((.(((.--.....)))----------..)--))...((((.(.(((-(((((-((((.......))))))..)))))).)((((....))))...))))... ( -21.90) >consensus CA_AUGAUAACCCCUUC____AA_AGGGGGU__UUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAU ..........((((...........))))..............(.(((.((((((((((.......))).)))))))))).)((((((......))).)))... (-14.17 = -15.25 + 1.08)


| Location | 7,224,779 – 7,224,895 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.28 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
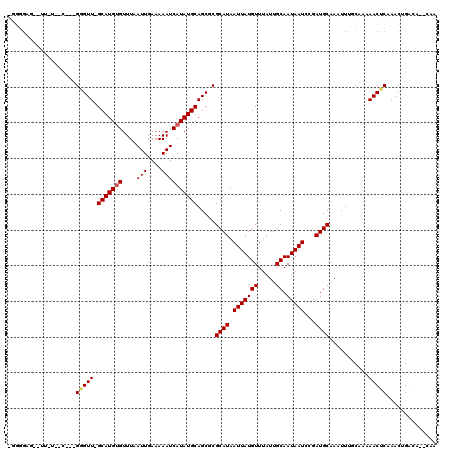
>X_DroMel_CAF1 7224779 116 + 22224390 AGGGGGU--UUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAUAAUCCGAUGCAAAUUUGCAAAAACUCAAACUGACA--CAA ((.((((--((((...((((.((((.((((((....(((....))))))))))))).)((((.(((((((.....)).)))))...)))).....)))))))))))...))....--... ( -26.20) >DroVir_CAF1 23838 100 + 1 UGGGGAG--UU--------GUGUU-GCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCUCGCAUAAUUAUGUUUAUUGCAAUAAUCUGAUGCGAAUUUUCAA-AACACAAACUG-------- ((..((.--..--------..(((-(((((((....(((....)))))))))))))((((((.(((((((.....)).)))))...)))))).))..)).-...........-------- ( -25.10) >DroPse_CAF1 1509 112 + 1 -----UG--CAGCAGCAGCGGGUU-GCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAUAAUCCGAUGCAAAUUUUCCAAAACUCCAACUGACACACAA -----((--(((.....(((.(((-(((((((....(((....))))))))))))).))).............(((((.........))))).................))).))..... ( -26.10) >DroYak_CAF1 1968 118 + 1 CGGGGGCCUUUUUCACGCAGGGUUGGCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAUAAUCCGAUGCAAAUUUGCAAAAACUCAAACUGACA--CAA .....(((...(((.....)))..))).((((((...((((.......(((((.....))))).....((((.((((((...............))))))))))))))...))))--)). ( -23.96) >DroMoj_CAF1 26811 100 + 1 UGGGGAG--UU--------GUGUU-GCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCUCGCAUAAUUAUGUUUAUUGCAAUAAUCUGAUGCAAAUUUUCAA-AACUCAAACUG-------- .((.(((--((--------(.(((-(((((((....(((....))))))))))))).)((((.(((((((.....)).)))))...))))..........-)))))...)).-------- ( -25.00) >DroAna_CAF1 1249 110 + 1 ----GCU--CCGUGGCAACGGGUU-GCAUG-GUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAUAAUCCGAUGCAAAUUCGCAAAAACUCAAACUGACA--CAA ----...--..((((((.((((((-(((((-((((.......))))))..))))))(((.((((......)))))))......))).)))..............((.....))))--).. ( -24.10) >consensus _GGGGAG__UU_U__C___GGGUU_GCAUGUGUUUAAUUGAAAAAUCAUAUGCAGCGCGCAUAAUUAUGUUUAUUGCAAUAAUCCGAUGCAAAUUUGCAAAAACUCAAACUGACA__CAA ...................(((((.((((((((((.......))).))))))).....((((.(((((((.....)).)))))...))))...........))))).............. (-16.17 = -16.28 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:11 2006