
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,214,751 – 7,214,851 |
| Length | 100 |
| Max. P | 0.656239 |

| Location | 7,214,751 – 7,214,851 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -21.79 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7214751 100 + 22224390 UGAAAGUGCUA-CCCUGGACCACUAAAAUAGUCCCCACGCCCC-----------------AACAGAAAUGAUAAUACAUUAUCAGGCCGGAGCAUUAAUGAGCUGAAGCUCAAACGCA ....((((((.-((..((((..........))))....(((..-----------------........((((((....))))))))).))))))))..(((((....)))))...... ( -23.30) >DroPse_CAF1 33732 111 + 1 UGAGAGUACUAUCCCUGGCAUACUAAAAUCCCCCCCACUGCUAAAAACGACGC-------CCCAGAAAUGAUAAUACAUUAUCAAGCCCAAGCAUUAAUGAGCUGAAGCUCAAACACA ...((((.........(((..................(((.............-------..)))...((((((....)))))).)))..(((........)))...))))....... ( -14.16) >DroSec_CAF1 22503 100 + 1 UGAAAGUGCUU-CCCUGGACCACUAAAAUAGUCCCCACGCCCC-----------------AACAGAAAUGAUAAUACAUUAUCAGGCCGGAGCAUUAAUGAGCUGAAGCUCAAACGCA ....(((((((-(...((((..........))))....(((..-----------------........((((((....))))))))).))))))))..(((((....)))))...... ( -24.20) >DroEre_CAF1 24375 100 + 1 UGAAAGUGCUA-CCCUGGACCACUAAAACAGUCCCCAUGCCCC-----------------AACUGAAAUGAUAAUACAUUAUCAGGCCGGAGCAUUAAUGAGCUGAAGCUCAAACGCA ....((((((.-..((((.((.......((((...........-----------------.))))...((((((....))))))))))))))))))..(((((....)))))...... ( -24.70) >DroYak_CAF1 25363 115 + 1 UGAAAGUGCUA-CCCUGGACCACUAAAAUAGUCCCCACGCCCC--CCCAACCCCCCAAAAAACUGAAAUGAUAAUACAUUAUCAGGCCGGAGCAUUAAUGAGCUGAAGCUCAAACGCA ....((((((.-((..((((..........))))....(((..--..........((......))...((((((....))))))))).))))))))..(((((....)))))...... ( -24.30) >DroAna_CAF1 24017 100 + 1 CGAAAGUGCUA-CCCUGGACCACUGAGAACUCACCCAUGCCCC-----------------GACACAAAUGAUAAUACAUUAUCAAGCCCGGGCAUUAAUGAGCUGGAGCUCAAACACA .....(((...-.(((((.....(((....)))..........-----------------........((((((....))))))...)))))......(((((....)))))..))). ( -20.10) >consensus UGAAAGUGCUA_CCCUGGACCACUAAAAUAGUCCCCACGCCCC_________________AACAGAAAUGAUAAUACAUUAUCAGGCCGGAGCAUUAAUGAGCUGAAGCUCAAACGCA ....((((((....((((.((.........((......))............................((((((....))))))))))))))))))..(((((....)))))...... (-15.02 = -15.67 + 0.64)

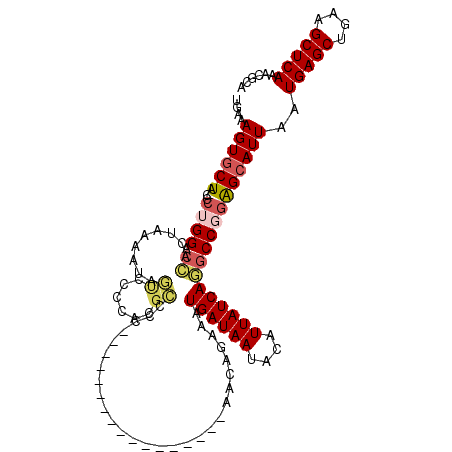

| Location | 7,214,751 – 7,214,851 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -34.13 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.09 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7214751 100 - 22224390 UGCGUUUGAGCUUCAGCUCAUUAAUGCUCCGGCCUGAUAAUGUAUUAUCAUUUCUGUU-----------------GGGGCGUGGGGACUAUUUUAGUGGUCCAGGG-UAGCACUUUCA (((...(((((....)))))...((((((((((..((.((((......)))))).)))-----------------)))))))..(((((((....)))))))....-..)))...... ( -34.30) >DroPse_CAF1 33732 111 - 1 UGUGUUUGAGCUUCAGCUCAUUAAUGCUUGGGCUUGAUAAUGUAUUAUCAUUUCUGGG-------GCGUCGUUUUUAGCAGUGGGGGGGAUUUUAGUAUGCCAGGGAUAGUACUCUCA .....(..((((..(((........)))..))))..)....((((((((...(((((.-------(((((.((((((....)))))).)))....))...)))))))))))))..... ( -27.00) >DroSec_CAF1 22503 100 - 1 UGCGUUUGAGCUUCAGCUCAUUAAUGCUCCGGCCUGAUAAUGUAUUAUCAUUUCUGUU-----------------GGGGCGUGGGGACUAUUUUAGUGGUCCAGGG-AAGCACUUUCA (((...(((((....)))))...((((((((((..((.((((......)))))).)))-----------------)))))))..(((((((....)))))))....-..)))...... ( -34.30) >DroEre_CAF1 24375 100 - 1 UGCGUUUGAGCUUCAGCUCAUUAAUGCUCCGGCCUGAUAAUGUAUUAUCAUUUCAGUU-----------------GGGGCAUGGGGACUGUUUUAGUGGUCCAGGG-UAGCACUUUCA (((...(((((....)))))...((((((((((.(((.((((......))))))))))-----------------)))))))..((((..(....)..))))....-..)))...... ( -35.00) >DroYak_CAF1 25363 115 - 1 UGCGUUUGAGCUUCAGCUCAUUAAUGCUCCGGCCUGAUAAUGUAUUAUCAUUUCAGUUUUUUGGGGGGUUGGG--GGGGCGUGGGGACUAUUUUAGUGGUCCAGGG-UAGCACUUUCA .((((((((((....)))))..))))).((((((((((((....))))))(..(((....)))..)))))))(--..((.((..(((((((....)))))))....-..)).))..). ( -39.40) >DroAna_CAF1 24017 100 - 1 UGUGUUUGAGCUCCAGCUCAUUAAUGCCCGGGCUUGAUAAUGUAUUAUCAUUUGUGUC-----------------GGGGCAUGGGUGAGUUCUCAGUGGUCCAGGG-UAGCACUUUCG .((((.(((((....)))))....((((((((((......((.....(((((..(((.-----------------...)))..))))).....))..))))).)))-))))))..... ( -34.80) >consensus UGCGUUUGAGCUUCAGCUCAUUAAUGCUCCGGCCUGAUAAUGUAUUAUCAUUUCUGUU_________________GGGGCGUGGGGACUAUUUUAGUGGUCCAGGG_UAGCACUUUCA (((...(((((....)))))...(((((((((..((((((....))))))..)).....................)))))))..(((((((....))))))).......)))...... (-20.08 = -20.09 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:04 2006