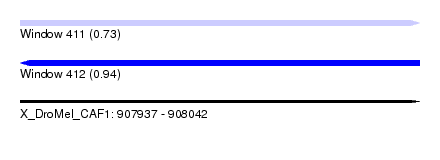
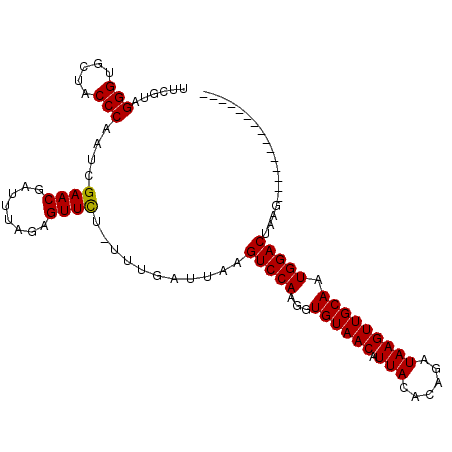
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 907,937 – 908,042 |
| Length | 105 |
| Max. P | 0.939297 |

| Location | 907,937 – 908,042 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.19 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 907937 105 + 22224390 UGCAACUUAACUGUGUUUGGUCCAUUGCAACUUAAGUGUGUAAUGUUACACAUUGGACUAAAUCAAA-AGAACUUUAUUUCGUUCGUUUGGGUAGCACCCUACUAA (((.((((((.((.((((((((((...........(((((((....)))))))))))))))))))..-.((((........))))..)))))).)))......... ( -26.40) >DroSec_CAF1 22816 90 + 1 ---------------CUGAGUCCAUUGCAACUUAUCUGUGUAAUGUUACACCUUGGACUUAAUCAAA-AGAACUCUAAAUCGUUCGAUUGGGUAGCACCCUACGAA ---------------.((((((((.((.(((((((....)))).))).))...)))))))).((((.-.((((........))))..))))((((....))))... ( -19.60) >DroSim_CAF1 21518 91 + 1 ---------------CUCAGUCCAUUGCAACUUAUCUGUGUAAUGUUACACCUUGGACUUAAUAAAAAAAAACUCUAAAUCGUUCGAUUGGGUAGCACCCUACGAA ---------------...((((((.((.(((((((....)))).))).))...)))))).......................((((...(((.....)))..)))) ( -15.70) >consensus _______________CUCAGUCCAUUGCAACUUAUCUGUGUAAUGUUACACCUUGGACUUAAUCAAA_AGAACUCUAAAUCGUUCGAUUGGGUAGCACCCUACGAA ................((((((((.............(((((....)))))..))))))))........((((........))))....(((.....)))...... (-15.97 = -16.19 + 0.23)

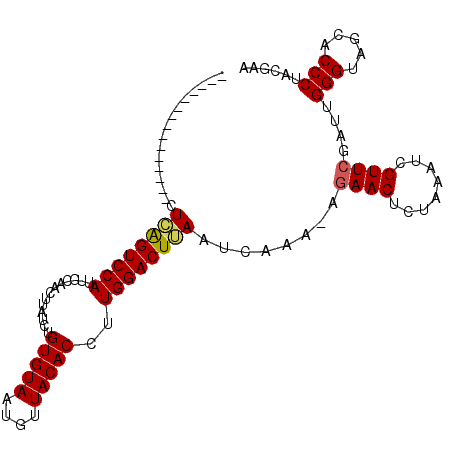
| Location | 907,937 – 908,042 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 907937 105 - 22224390 UUAGUAGGGUGCUACCCAAACGAACGAAAUAAAGUUCU-UUUGAUUUAGUCCAAUGUGUAACAUUACACACUUAAGUUGCAAUGGACCAAACACAGUUAAGUUGCA ...((((((.....))).(((((((........)))).-..((.(((.(((((...((((((.(((......))))))))).))))).)))))..)))....))). ( -23.00) >DroSec_CAF1 22816 90 - 1 UUCGUAGGGUGCUACCCAAUCGAACGAUUUAGAGUUCU-UUUGAUUAAGUCCAAGGUGUAACAUUACACAGAUAAGUUGCAAUGGACUCAG--------------- ......(((.....)))(((((((.((........)).-))))))).((((((...((((((.(((......))))))))).))))))...--------------- ( -23.00) >DroSim_CAF1 21518 91 - 1 UUCGUAGGGUGCUACCCAAUCGAACGAUUUAGAGUUUUUUUUUAUUAAGUCCAAGGUGUAACAUUACACAGAUAAGUUGCAAUGGACUGAG--------------- ((((..(((.....)))...)))).......................((((((...((((((.(((......))))))))).))))))...--------------- ( -20.50) >consensus UUCGUAGGGUGCUACCCAAUCGAACGAUUUAGAGUUCU_UUUGAUUAAGUCCAAGGUGUAACAUUACACAGAUAAGUUGCAAUGGACUAAG_______________ ......(((.....)))....((((........))))...........(((((...((((((.(((......))))))))).)))))................... (-19.92 = -19.70 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:29 2006