| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,201,731 – 7,201,862 |
| Length | 131 |
| Max. P | 0.997553 |

| Location | 7,201,731 – 7,201,844 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
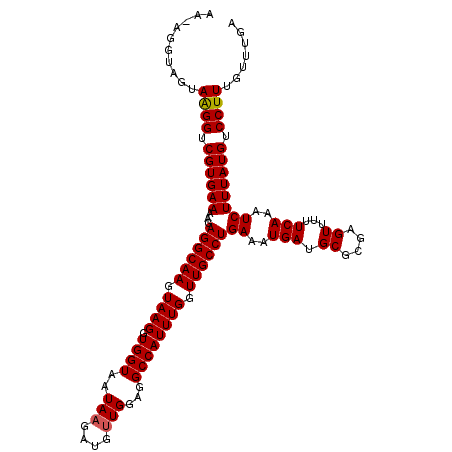
>X_DroMel_CAF1 7201731 113 - 22224390 AA-AGGCAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGCGCGAGUUUUUCAAAUCUUUAUGUCCUUUGUUUGA ((-(((..((((((...((((((.((((((.((((.((((..(((....)))...)))))))).))))))........((....))))))))...))))))..)))))...... ( -31.20) >DroSec_CAF1 9603 113 - 1 AA-AGGUAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGGUGGAGGCCAUUUGGUUGCCUGACAUGAUGCGCGAGUUUUUCAAAUCUUUAUGUCCUUUGUUUGA ((-(((..((((((...(((((((.((((........((((((.(((((((....))))))).))))))........))).)...)))))))...))))))..)))))...... ( -32.49) >DroSim_CAF1 9731 113 - 1 AA-AGGUAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGCGCGAGUUUUUCAAAUCUUUAUGUCCUUUGUUUGA ((-(((..((((((...((((((.((((((.((((.((((..(((....)))...)))))))).))))))........((....))))))))...))))))..)))))...... ( -31.20) >DroEre_CAF1 9412 113 - 1 AG-AGGUAGUAGGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGCGCGAGUUUUUCAAAUCUUUAUGUCCUUUGUUUGA ((-(((..((((((...((((((.((((((.((((.((((..(((....)))...)))))))).))))))........((....))))))))...))))))..)))))...... ( -30.90) >DroYak_CAF1 11364 114 - 1 AGGCGGUGGCAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUGAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGCGCGAGUUUUUCACAUCUUUAUGUCCUUUGUUUGA ((((((.((((((((.(((((((.((((((.((((.((((..(((....)))...)))))))).))))))........((....)))))))))))))...))))..)))))).. ( -30.20) >consensus AA_AGGUAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGCGCGAGUUUUUCAAAUCUUUAUGUCCUUUGUUUGA ..........((((.((((((...((((((.((((.((((..(((....)))...)))))))).))))))((..(((.((....))...)))..)))))))).))))....... (-28.20 = -28.24 + 0.04)


| Location | 7,201,765 – 7,201,862 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -16.39 |
| Consensus MFE | -13.13 |
| Energy contribution | -13.13 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
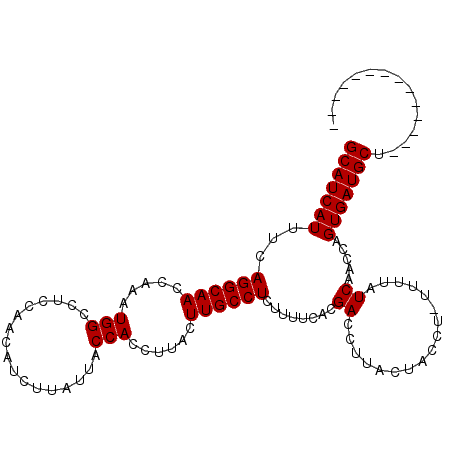
>X_DroMel_CAF1 7201765 97 + 22224390 GCAUCAUUUCAGGCAACCAAAUGGCCUCCAACAUCUUAUUACCACCUUACUUGCCUCUUUUCACGACCUUACUGCCU-UUUCAUCAACCAGUGAUGCU-------------- (((((.....((((((.....(((.................)))......))))))..............((((...-..........))))))))).-------------- ( -14.65) >DroSec_CAF1 9637 97 + 1 GCAUCAUGUCAGGCAACCAAAUGGCCUCCACCAUCUUAUUACCACCUUACUUGCCUCUUUUCACGACCUUACUACCU-UUUUAUCAACCAGUGAUGCU-------------- (((((((...((((((.....(((.................)))......))))))........((...........-.....)).....))))))).-------------- ( -13.72) >DroYak_CAF1 11398 112 + 1 GCAUCAUUUCAGGCAACCAAAUGGCCUCCAACAUCUUAUCACCACCUUACUUGCCUCUUUUCACGACCUUGCCACCGCCUUUAUCAACCAGUGAUGCCUGAAUGGUGAUAAC .(((((((.((((((.......(((...........................))).....((((..........................)))))))))))))))))..... ( -20.80) >consensus GCAUCAUUUCAGGCAACCAAAUGGCCUCCAACAUCUUAUUACCACCUUACUUGCCUCUUUUCACGACCUUACUACCU_UUUUAUCAACCAGUGAUGCU______________ (((((((...((((((.....(((.................)))......))))))........((.................)).....)))))))............... (-13.13 = -13.13 + -0.00)

| Location | 7,201,765 – 7,201,862 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.42 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
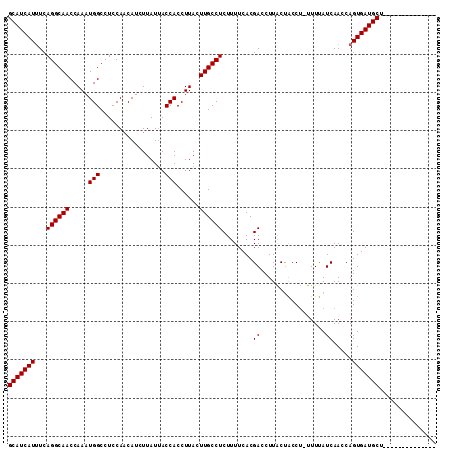
>X_DroMel_CAF1 7201765 97 - 22224390 --------------AGCAUCACUGGUUGAUGAAA-AGGCAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGC --------------.(((((((((.((.......-)).))))..............((((((.((((.((((..(((....)))...)))))))).)))))).....))))) ( -26.10) >DroSec_CAF1 9637 97 - 1 --------------AGCAUCACUGGUUGAUAAAA-AGGUAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGGUGGAGGCCAUUUGGUUGCCUGACAUGAUGC --------------.(((((((((.((.......-)).))))...(((((........)).........((((((.(((((((....))))))).)))))).)))..))))) ( -25.40) >DroYak_CAF1 11398 112 - 1 GUUAUCACCAUUCAGGCAUCACUGGUUGAUAAAGGCGGUGGCAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUGAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGC .(((((((((((...((.((((((.((.....)).))))))....(((.........)))..))..)))))))))))......((..(((....)))..))........... ( -30.30) >consensus ______________AGCAUCACUGGUUGAUAAAA_AGGUAGUAAGGUCGUGAAAAGAGGCAAGUAAGGUGGUAAUAAGAUGUUGGAGGCCAUUUGGUUGCCUGAAAUGAUGC ...............(((((((((.((........)).)))...............((((((.((((.((((..((......))...)))))))).))))))....)))))) (-21.34 = -21.23 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:55 2006