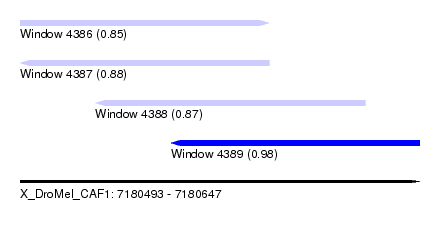
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 7,180,493 – 7,180,647 |
| Length | 154 |
| Max. P | 0.984158 |

| Location | 7,180,493 – 7,180,589 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 61.41 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -9.47 |
| Energy contribution | -9.40 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7180493 96 + 22224390 -------UGUUUAUUUGUUUGCAGGUGCAGCAGCAAAUAGAUUA-----UAAUUUAUGUAUUUGUAGCGGGCGCAUGCUAAACUGCACACACUCUACACCUACAUAUG -------.((((((((((((((....)))..)))))))))))..-----.....((((((..(((((.(.(.(((........))).)...).)))))..)))))).. ( -24.30) >DroSec_CAF1 5954 86 + 1 -------UGUUUAUUUGUUUGCAGGUGCAGCAGCAAAUAGAUUA-----UAAUUUAUGUAUUUGUAGCGGGCGCAUGCUAAACUACACACACUCUA----------UA -------.........(((((((.((((.((.(((((((.((..-----......)).))))))).))..)))).))).)))).............----------.. ( -22.60) >DroEre_CAF1 7662 105 + 1 CACCUUCUGUUUGGU--UUUGCAGGUGCAGCAGCAAAUAGAUUGUAUGUAAUUGUGUGUAUUUGUAGCGGGCGCAUGCUAAACUGCAUACACCCUAUACAUACAUAC- .((..((((((((.(--.((((....)))).).))))))))..))........((((((((.(((((.((..(.((((......)))).).))))))).))))))))- ( -33.80) >DroAna_CAF1 12296 74 + 1 -------UGUGU------CUUGAGAUACA-------------CA-----GGGCUGCUGUAUUUUUAGCAGCCAUAUGUUAAA-UGCAC-CACCCCAGAAAUCCAUAC- -------.((((------(....)))))(-------------((-----.((((((((......))))))))...)))....-.....-..................- ( -20.90) >consensus _______UGUUUAUU__UUUGCAGGUGCAGCAGCAAAUAGAUUA_____UAAUUUAUGUAUUUGUAGCGGGCGCAUGCUAAACUGCACACACCCUA_ACAUACAUAC_ ..................(((((((((((...........................))))))))))).(((.(..(((......)))..).))).............. ( -9.47 = -9.40 + -0.06)


| Location | 7,180,493 – 7,180,589 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 61.41 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7180493 96 - 22224390 CAUAUGUAGGUGUAGAGUGUGUGCAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA-----UAAUCUAUUUGCUGCUGCACCUGCAAACAAAUAAACA------- ....(((((((((((.((((((((......)))))))).)((..((((((.((......-----..)).))))))..))))))))))))............------- ( -31.10) >DroSec_CAF1 5954 86 - 1 UA----------UAGAGUGUGUGUAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA-----UAAUCUAUUUGCUGCUGCACCUGCAAACAAAUAAACA------- ..----------....((((.((((((...((....))..)))))).))))........-----.....((((((.(((.......)))..))))))....------- ( -16.50) >DroEre_CAF1 7662 105 - 1 -GUAUGUAUGUAUAGGGUGUAUGCAGUUUAGCAUGCGCCCGCUACAAAUACACACAAUUACAUACAAUCUAUUUGCUGCUGCACCUGCAAA--ACCAAACAGAAGGUG -(((((((((((..((((((((((......))))))))))..))))............)))))))..(((.((((.(..(((....)))..--).)))).)))..... ( -32.70) >DroAna_CAF1 12296 74 - 1 -GUAUGGAUUUCUGGGGUG-GUGCA-UUUAACAUAUGGCUGCUAAAAAUACAGCAGCCC-----UG-------------UGUAUCUCAAG------ACACA------- -...((..(((.((((((.-.....-....(((((.(((((((........))))))).-----))-------------)))))))))))------)..))------- ( -20.40) >consensus _AUAUGUAUGU_UAGAGUGUGUGCAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA_____UAAUCUAUUUGCUGCUGCACCUGCAAA__AAUAAACA_______ ..............((((((((((......)))))))))).................................................................... ( -9.24 = -9.80 + 0.56)


| Location | 7,180,522 – 7,180,626 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 57.68 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7180522 104 - 22224390 UCUCCAACCAUGUAGUAUAU---ACAUACAUAUGUAUGUACAUAUGUAGGUGUAGAGUGUGUGCAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA-----UAAUCUAU ..........((((((((((---((((....)))))))))).((((((..(((((.((((((((......))))))))...)))))..))))))...))-----))...... ( -34.20) >DroSec_CAF1 5983 85 - 1 UCUC-AACCAUGUAGUAUAU-----------AUGUAUGUAUA----------UAGAGUGUGUGUAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA-----UAAUCUAU ....-......((((..(((-----------(..(((((((.----------...((((.(((((........))))).))))....)))))))...))-----))..)))) ( -19.10) >DroEre_CAF1 7696 105 - 1 UCGC-UACCACAUAGUAUAUCGCACAUACAAGUGU------GUAUGUAUGUAUAGGGUGUAUGCAGUUUAGCAUGCGCCCGCUACAAAUACACACAAUUACAUACAAUCUAU ..((-((.....))))...............((((------((((...((((..((((((((((......))))))))))..)))).))))))))................. ( -33.60) >DroAna_CAF1 12312 78 - 1 UCUU-GGCCC--------------CAUGCAAGUCU------GUAUGGAUUUCUGGGGUG-GUGCA-UUUAACAUAUGGCUGCUAAAAAUACAGCAGCCC-----UG------ ...(-.((((--------------((.(.((((((------....))))))))))))).-)....-..........(((((((........))))))).-----..------ ( -26.50) >consensus UCUC_AACCAUGUAGUAUAU____CAUACAAAUGU______AUAUGUAUGU_UAGAGUGUGUGCAGUUUAGCAUGCGCCCGCUACAAAUACAUAAAUUA_____UAAUCUAU ......................................................((((((((((......))))))))))................................ ( -9.24 = -9.80 + 0.56)


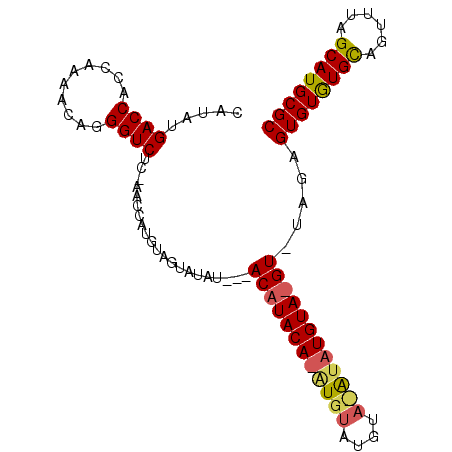
| Location | 7,180,551 – 7,180,647 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 73.38 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -15.68 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 7180551 96 - 22224390 CAUAUGACCACCAAAACAGGGUCUCCAACCAUGUAGUAUAU---ACAUACAUAUGUAUGUACAUAUGUAGGUGUAGAGUGUGUGCAGUUUAGCAUGCGC ........((((...(((.(((.....))).))).((((((---((((....)))))))))).......))))....((((((((......)))))))) ( -29.40) >DroSec_CAF1 6012 77 - 1 CAUAUGACCACCAAAACAGGGUCUC-AACCAUGUAGUAUAU-----------AUGUAUGUAUA----------UAGAGUGUGUGUAGUUUAGCAUGCGC .....((((..........))))..-..........(((((-----------((....)))))----------))..((((((((......)))))))) ( -14.50) >DroEre_CAF1 7730 92 - 1 CAUAUGACCACCAAAACAGGGUCGC-UACCACAUAGUAUAUCGCACAUACAAGUGU------GUAUGUAUGUAUAGGGUGUAUGCAGUUUAGCAUGCGC .....((((..........)))).(-((..(((((.((((.(((((......))))------)))))))))).))).((((((((......)))))))) ( -26.70) >consensus CAUAUGACCACCAAAACAGGGUCUC_AACCAUGUAGUAUAU___ACAUACA_AUGUAUGUA_AUAUGUA_GU_UAGAGUGUGUGCAGUUUAGCAUGCGC .....((((..........)))).....................((((((((((((....)))))))))))).....((((((((......)))))))) (-15.68 = -17.13 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:47 2006